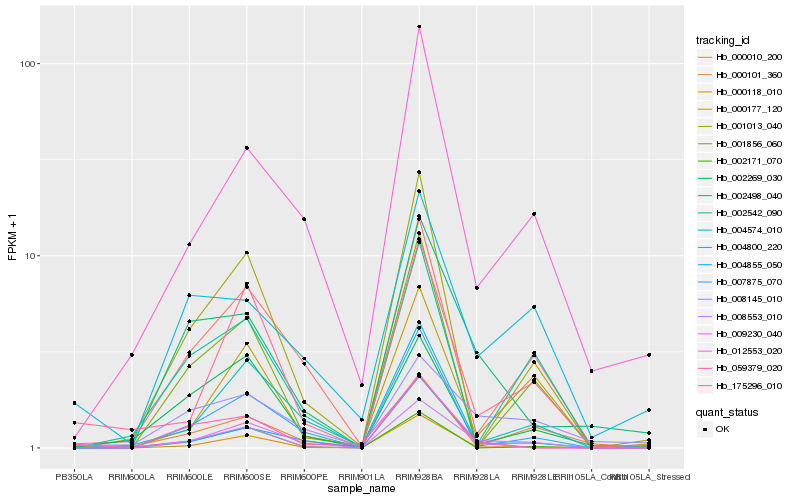
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008553_010 |
0.0 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 2 |
Hb_175296_010 |
0.1643060776 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 3 |
Hb_000118_010 |
0.2056284841 |
desease resistance |
Gene Name: LRR_4 |
hypothetical protein POPTR_0021s00470g [Populus trichocarpa] |
| 4 |
Hb_000101_360 |
0.2189293434 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 5 |
Hb_002498_040 |
0.2221376231 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 6 |
Hb_012553_020 |
0.2253760595 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 7 |
Hb_000010_200 |
0.225834823 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001013_040 |
0.2258404304 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 9 |
Hb_000177_120 |
0.2261425995 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008145_010 |
0.2269786939 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_004855_050 |
0.2282729772 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 12 |
Hb_004800_220 |
0.2310216196 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 13 |
Hb_059379_020 |
0.2316465963 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase 1-like [Populus euphratica] |
| 14 |
Hb_001856_060 |
0.2317834876 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_004574_010 |
0.2334771401 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 16 |
Hb_007875_070 |
0.2345051753 |
- |
- |
hypothetical protein JCGZ_05017 [Jatropha curcas] |
| 17 |
Hb_002542_090 |
0.2369022903 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 18 |
Hb_009230_040 |
0.2391671236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002269_030 |
0.2438157647 |
- |
- |
hypothetical protein CISIN_1g018400mg [Citrus sinensis] |
| 20 |
Hb_002171_070 |
0.2443813277 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance gene-like protein ARGH30 [Populus trichocarpa] |