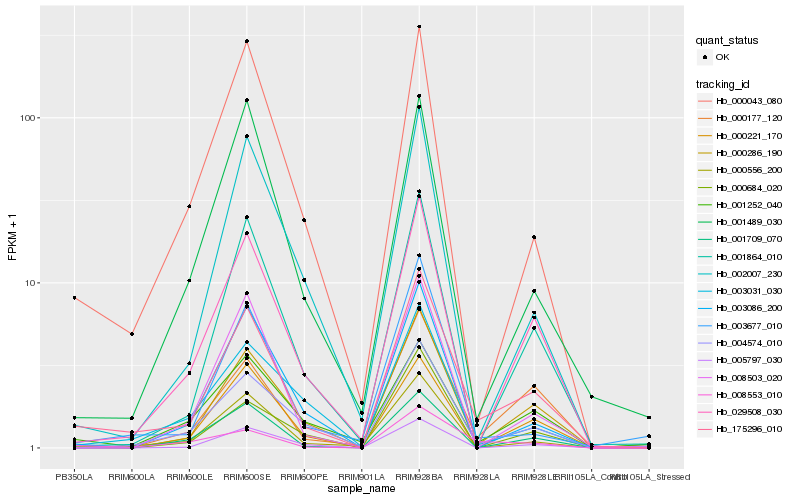
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_175296_010 |
0.0 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 2 |
Hb_029508_030 |
0.1591057536 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0002s04880g [Populus trichocarpa] |
| 3 |
Hb_004574_010 |
0.1625843616 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 4 |
Hb_008553_010 |
0.1643060776 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 5 |
Hb_000286_190 |
0.1649309444 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 6 |
Hb_000043_080 |
0.167859537 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 7 |
Hb_001864_010 |
0.1691498057 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000177_120 |
0.1727672661 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003677_010 |
0.1757794878 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 10 |
Hb_000556_200 |
0.1774662544 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 22 [Jatropha curcas] |
| 11 |
Hb_003086_200 |
0.180671906 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 12 |
Hb_002007_230 |
0.1823690352 |
- |
- |
PREDICTED: probable linoleate 9S-lipoxygenase 5 [Jatropha curcas] |
| 13 |
Hb_000221_170 |
0.1831148601 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 2 [Jatropha curcas] |
| 14 |
Hb_005797_030 |
0.188318005 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like [Jatropha curcas] |
| 15 |
Hb_003031_030 |
0.1883915905 |
- |
- |
hypothetical protein JCGZ_21288 [Jatropha curcas] |
| 16 |
Hb_008503_020 |
0.1884010806 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 17 |
Hb_001709_070 |
0.1894951658 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 18 |
Hb_001489_030 |
0.189634892 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 19 |
Hb_000684_020 |
0.1902259968 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_001252_040 |
0.1902671643 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |