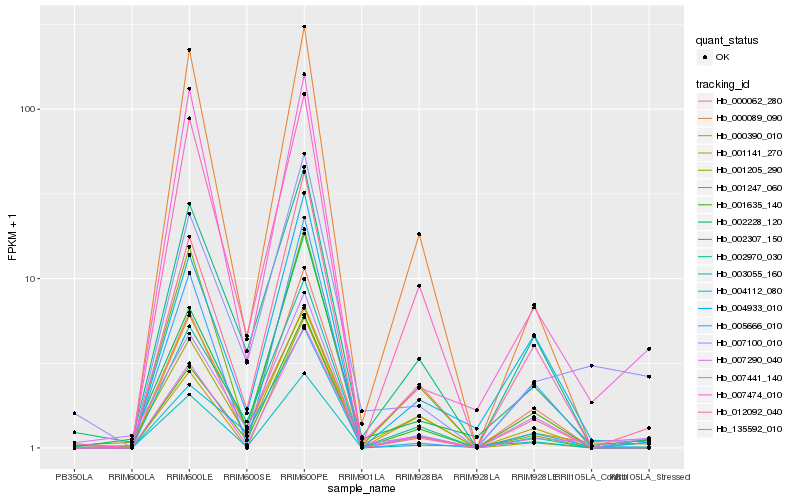
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_140 |
0.0 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 2 |
Hb_007100_010 |
0.1161437534 |
- |
- |
hypothetical protein ARALYDRAFT_479822 [Arabidopsis lyrata subsp. lyrata] |
| 3 |
Hb_007474_010 |
0.1517710961 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 4 |
Hb_000089_090 |
0.1577586574 |
- |
- |
hypothetical protein POPTR_0019s12670g [Populus trichocarpa] |
| 5 |
Hb_135592_010 |
0.1604564883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002228_120 |
0.1627304861 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 7 |
Hb_002970_030 |
0.1630520012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_012092_040 |
0.1654433047 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003055_160 |
0.1656102627 |
- |
- |
PREDICTED: S-linalool synthase isoform X1 [Jatropha curcas] |
| 10 |
Hb_000390_010 |
0.1661962968 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 11 |
Hb_005666_010 |
0.16816594 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 12 |
Hb_001635_140 |
0.169047602 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_004112_080 |
0.169280803 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 14 |
Hb_001141_270 |
0.1702722093 |
- |
- |
PREDICTED: uncharacterized protein LOC105632005 [Jatropha curcas] |
| 15 |
Hb_002307_150 |
0.1703007369 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 16 |
Hb_007290_040 |
0.1708815423 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1-like [Jatropha curcas] |
| 17 |
Hb_001247_060 |
0.1737926502 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000062_280 |
0.1758742962 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 19 |
Hb_004933_010 |
0.1759029785 |
- |
- |
hypothetical protein POPTR_0018s10810g [Populus trichocarpa] |
| 20 |
Hb_001205_290 |
0.176479669 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |