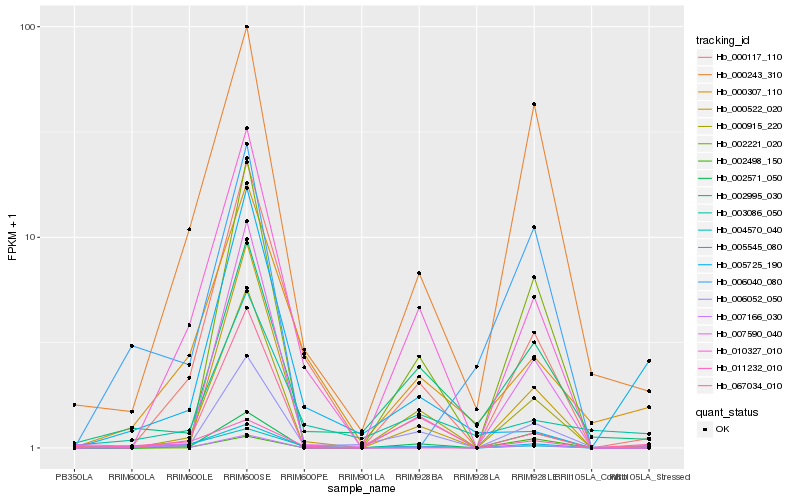
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007166_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_011232_010 |
0.2600969031 |
- |
- |
Leucine-rich repeat receptor-like protein kinase family protein [Theobroma cacao] |
| 3 |
Hb_002995_030 |
0.262826333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000915_220 |
0.2655916267 |
- |
- |
Lactoylglutathione lyase / glyoxalase I family protein [Theobroma cacao] |
| 5 |
Hb_004570_040 |
0.2667075301 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_007590_040 |
0.2776187357 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like [Jatropha curcas] |
| 7 |
Hb_002571_050 |
0.2814523203 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 8 |
Hb_005545_080 |
0.2826495908 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |
| 9 |
Hb_002221_020 |
0.2863580021 |
- |
- |
hypothetical protein B456_008G083700 [Gossypium raimondii] |
| 10 |
Hb_000522_020 |
0.2867564522 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 11 |
Hb_000243_310 |
0.288112064 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 12 |
Hb_067034_010 |
0.288508229 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like [Jatropha curcas] |
| 13 |
Hb_006052_050 |
0.2917859431 |
- |
- |
hypothetical protein EUGRSUZ_A00568 [Eucalyptus grandis] |
| 14 |
Hb_006040_080 |
0.2934162225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000307_110 |
0.2961506811 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 16 |
Hb_005725_190 |
0.296332491 |
- |
- |
hypothetical protein JCGZ_15992 [Jatropha curcas] |
| 17 |
Hb_000117_110 |
0.2994278561 |
- |
- |
transferase, putative [Ricinus communis] |
| 18 |
Hb_002498_150 |
0.301274952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003086_050 |
0.3026403682 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 20 |
Hb_010327_010 |
0.3034170446 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |