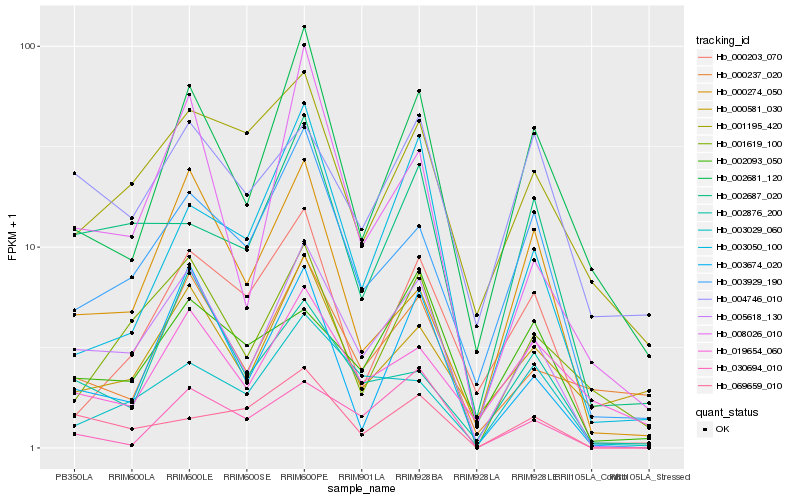
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005618_130 |
0.0 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 2 |
Hb_003674_020 |
0.167875199 |
- |
- |
PREDICTED: DNA replication licensing factor MCM6 [Jatropha curcas] |
| 3 |
Hb_003929_190 |
0.172890505 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 4 |
Hb_008026_010 |
0.1747922198 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 9 [Jatropha curcas] |
| 5 |
Hb_002681_120 |
0.1762928572 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 6 |
Hb_002687_020 |
0.182479129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001619_100 |
0.1841526513 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 8 |
Hb_003029_060 |
0.199067256 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000237_020 |
0.2001075686 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 10 |
Hb_001195_420 |
0.2065060719 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 11 |
Hb_069659_010 |
0.2065796883 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 12 |
Hb_002093_050 |
0.2068146245 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 13 |
Hb_000274_050 |
0.2069414737 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 14 |
Hb_030694_010 |
0.2071453118 |
- |
- |
PREDICTED: endoglucanase 25-like [Solanum tuberosum] |
| 15 |
Hb_002876_200 |
0.2075145633 |
- |
- |
PREDICTED: uncharacterized protein LOC105641209 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000203_070 |
0.207991987 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 17 |
Hb_000581_030 |
0.2092498865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004746_010 |
0.213564855 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 19 |
Hb_019654_060 |
0.2142625274 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_003050_100 |
0.2170019281 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |