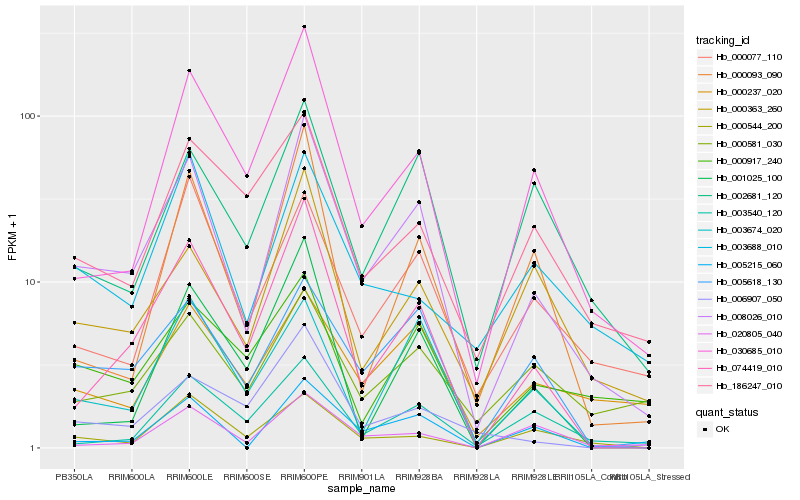
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008026_010 |
0.0 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 9 [Jatropha curcas] |
| 2 |
Hb_074419_010 |
0.1521634158 |
- |
- |
PREDICTED: uncharacterized protein LOC105637596 [Jatropha curcas] |
| 3 |
Hb_005618_130 |
0.1747922198 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 4 |
Hb_030685_010 |
0.1749534145 |
- |
- |
- |
| 5 |
Hb_005215_060 |
0.1911071409 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 6 |
Hb_000363_260 |
0.1919798563 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 7 |
Hb_002681_120 |
0.1935366887 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 8 |
Hb_000093_090 |
0.1972387962 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 9 |
Hb_003674_020 |
0.1977241032 |
- |
- |
PREDICTED: DNA replication licensing factor MCM6 [Jatropha curcas] |
| 10 |
Hb_000077_110 |
0.1997323129 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |
| 11 |
Hb_000237_020 |
0.2029276104 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 12 |
Hb_000544_200 |
0.2057513006 |
- |
- |
PREDICTED: uncharacterized protein LOC105647171 [Jatropha curcas] |
| 13 |
Hb_000581_030 |
0.2090844846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006907_050 |
0.2092556657 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_003540_120 |
0.2096402448 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 16 |
Hb_001025_100 |
0.2109205167 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000917_240 |
0.2114234115 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 18 |
Hb_020805_040 |
0.2125894856 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1A [Jatropha curcas] |
| 19 |
Hb_003688_010 |
0.2150265895 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 20 |
Hb_186247_010 |
0.2163072291 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |