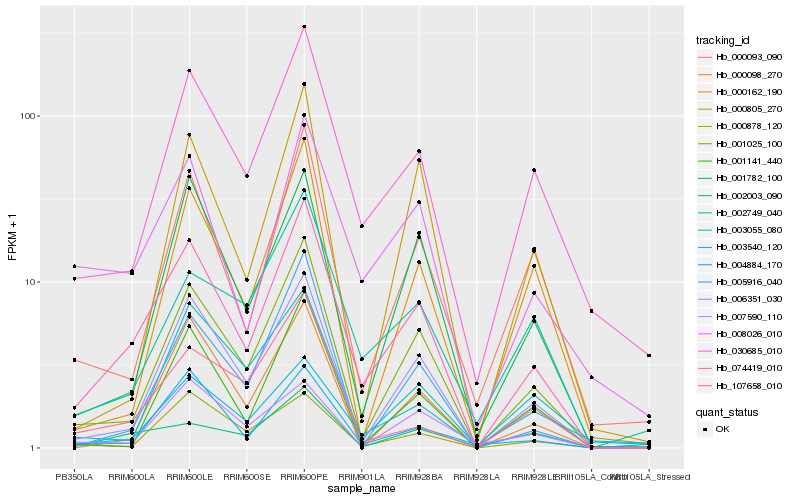
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074419_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637596 [Jatropha curcas] |
| 2 |
Hb_001025_100 |
0.1241700375 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 3 |
Hb_006351_030 |
0.1364394359 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MARCH8-like [Jatropha curcas] |
| 4 |
Hb_030685_010 |
0.1400122904 |
- |
- |
- |
| 5 |
Hb_008026_010 |
0.1521634158 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 9 [Jatropha curcas] |
| 6 |
Hb_000093_090 |
0.173143016 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 7 |
Hb_001782_100 |
0.1749356165 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 8 |
Hb_005916_040 |
0.1805014944 |
- |
- |
PREDICTED: auxin transporter-like protein 5 [Populus euphratica] |
| 9 |
Hb_002003_090 |
0.1836466906 |
- |
- |
PREDICTED: TPR repeat-containing protein ZIP4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000098_270 |
0.1860461035 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 11 |
Hb_002749_040 |
0.1889848559 |
- |
- |
Ribosomal protein L6 family protein [Theobroma cacao] |
| 12 |
Hb_004884_170 |
0.1891742995 |
- |
- |
hypothetical protein AALP_AA1G067000 [Arabis alpina] |
| 13 |
Hb_001141_440 |
0.1895608519 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 14 |
Hb_107658_010 |
0.1911492837 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000878_120 |
0.1913570451 |
- |
- |
PREDICTED: protein downstream neighbor of Son [Jatropha curcas] |
| 16 |
Hb_003055_080 |
0.1956834849 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000162_190 |
0.1960278415 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 18 |
Hb_000805_270 |
0.1987112082 |
- |
- |
PREDICTED: expansin-A4 [Gossypium raimondii] |
| 19 |
Hb_003540_120 |
0.1993863639 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_007590_110 |
0.2000398361 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |