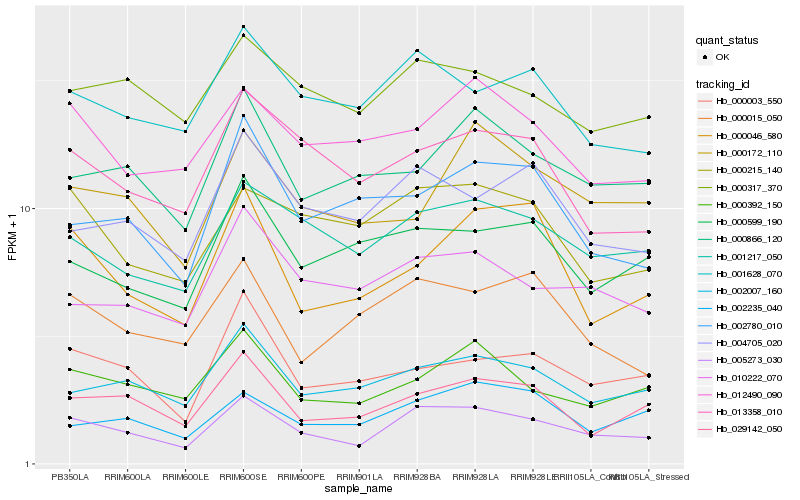
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005273_030 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 2 |
Hb_001217_050 |
0.1128720704 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 3 |
Hb_002007_160 |
0.1173075645 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 4 |
Hb_029142_050 |
0.1176444998 |
desease resistance |
Gene Name: PPR |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 5 |
Hb_000392_150 |
0.1178148168 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g74580 [Jatropha curcas] |
| 6 |
Hb_001628_070 |
0.1216486341 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_000046_580 |
0.1225097281 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 8 |
Hb_013358_010 |
0.1229042424 |
- |
- |
hypothetical protein JCGZ_07102 [Jatropha curcas] |
| 9 |
Hb_004705_020 |
0.1236681985 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 10 |
Hb_000215_140 |
0.1237626174 |
- |
- |
PREDICTED: phosphoinositide 3-kinase regulatory subunit 4-like [Malus domestica] |
| 11 |
Hb_010222_070 |
0.126223319 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000003_550 |
0.1269571237 |
- |
- |
- |
| 13 |
Hb_000866_120 |
0.1275379682 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 14 |
Hb_000015_050 |
0.1283617367 |
- |
- |
hypothetical protein JCGZ_01495 [Jatropha curcas] |
| 15 |
Hb_000317_370 |
0.1302417377 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 16 |
Hb_012490_090 |
0.1303342881 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 17 |
Hb_002235_040 |
0.1306063488 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 18 |
Hb_000172_110 |
0.1326242002 |
- |
- |
- |
| 19 |
Hb_002780_010 |
0.1330189188 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 20 |
Hb_000599_190 |
0.1332782523 |
transcription factor |
TF Family: SNF2 |
DNA repair and recombination protein RAD26, putative [Ricinus communis] |