| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
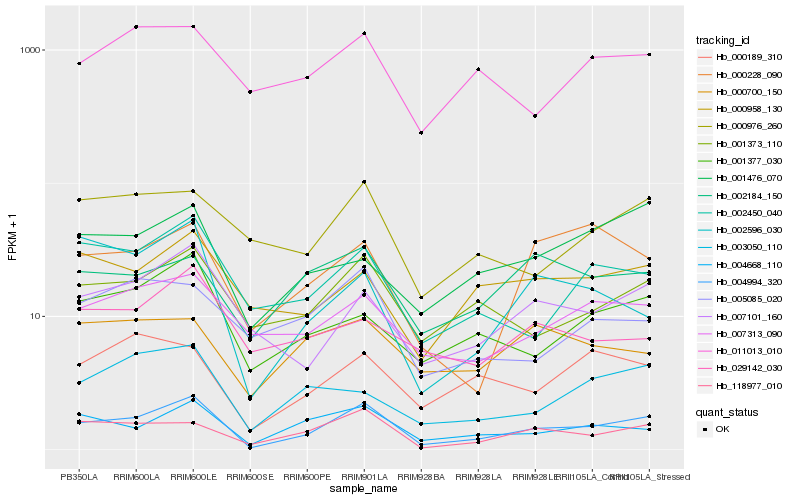
Hb_004994_320 |
0.0 |
- |
- |
PREDICTED: putative ripening-related protein 1 [Cicer arietinum] |
| 2 |
Hb_118977_010 |
0.1615846558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004668_110 |
0.1655307463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001373_110 |
0.1720672681 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 5 |
Hb_007101_160 |
0.1769334756 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001377_030 |
0.1822891586 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 7 |
Hb_002450_040 |
0.1843873655 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 6B [Vitis vinifera] |
| 8 |
Hb_007313_090 |
0.1899595715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000976_260 |
0.1978245325 |
- |
- |
- |
| 10 |
Hb_000189_310 |
0.1983087822 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003050_110 |
0.1989031214 |
- |
- |
PREDICTED: prefoldin subunit 6 [Populus euphratica] |
| 12 |
Hb_000958_130 |
0.2002391091 |
- |
- |
PREDICTED: uncharacterized protein LOC105628716 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002184_150 |
0.2008771519 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_002596_030 |
0.2033128812 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase [NAD(+)] GPDHC1, cytosolic [Jatropha curcas] |
| 15 |
Hb_001476_070 |
0.2037063017 |
- |
- |
MinE protein [Manihot esculenta] |
| 16 |
Hb_005085_020 |
0.2048636027 |
- |
- |
hypothetical protein B456_001G193200 [Gossypium raimondii] |
| 17 |
Hb_000228_090 |
0.2053168069 |
- |
- |
PREDICTED: protein PAM68, chloroplastic [Jatropha curcas] |
| 18 |
Hb_011013_010 |
0.2078304174 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 19 |
Hb_000700_150 |
0.2114625754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_029142_030 |
0.2138397791 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |