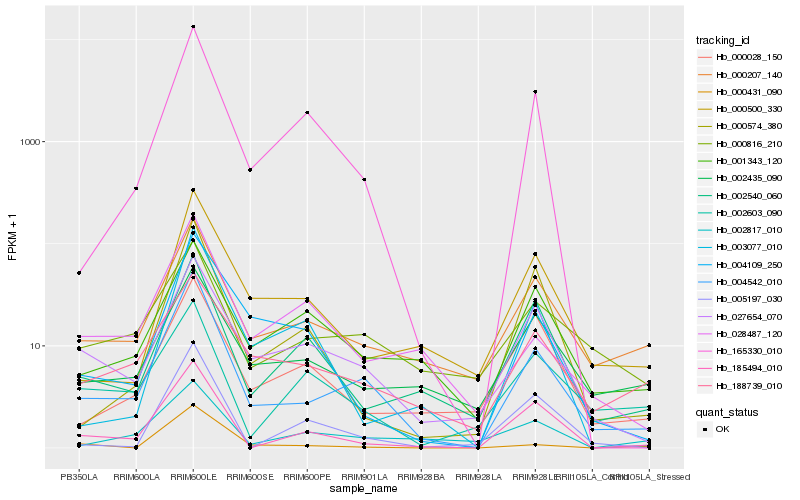
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004542_010 |
0.0 |
- |
- |
Putative chloroplast RF19 [Medicago truncatula] |
| 2 |
Hb_000207_140 |
0.1838253559 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 3 |
Hb_027654_070 |
0.1935295734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_185494_010 |
0.1943264924 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000500_330 |
0.202299918 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 6 |
Hb_165330_010 |
0.2064289606 |
- |
- |
BnaUnng00840D [Brassica napus] |
| 7 |
Hb_004109_250 |
0.2171252985 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 8 |
Hb_188739_010 |
0.2182492079 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_002540_060 |
0.2209582073 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000816_210 |
0.2210169297 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase isoform X2 [Jatropha curcas] |
| 11 |
Hb_000574_380 |
0.2228361508 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 12 |
Hb_028487_120 |
0.2229406918 |
- |
- |
PREDICTED: HMG-Y-related protein B-like [Jatropha curcas] |
| 13 |
Hb_005197_030 |
0.2241506012 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH71-like isoform X2 [Populus euphratica] |
| 14 |
Hb_002435_090 |
0.2256049669 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000431_090 |
0.2261353391 |
- |
- |
hypothetical protein POPTR_0017s14020g [Populus trichocarpa] |
| 16 |
Hb_001343_120 |
0.2325455183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003077_010 |
0.2354773436 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000028_150 |
0.2385091428 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 19 |
Hb_002817_010 |
0.239892579 |
- |
- |
PREDICTED: uncharacterized protein LOC105630103 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002603_090 |
0.2401445527 |
- |
- |
PREDICTED: GDSL esterase/lipase CPRD49-like isoform X2 [Populus euphratica] |