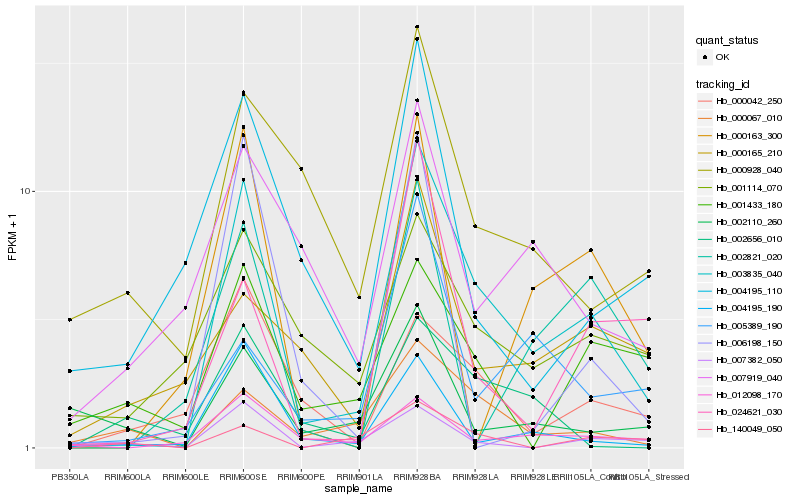
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003835_040 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 91A1-like [Jatropha curcas] |
| 2 |
Hb_012098_170 |
0.2425684829 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_16874 [Jatropha curcas] |
| 3 |
Hb_004195_190 |
0.2583195832 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_000067_010 |
0.259135985 |
- |
- |
hypothetical protein B456_002G147500 [Gossypium raimondii] |
| 5 |
Hb_007382_050 |
0.2737310177 |
- |
- |
PREDICTED: uncharacterized protein LOC105645009 [Jatropha curcas] |
| 6 |
Hb_000163_300 |
0.273949665 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 7 |
Hb_000042_250 |
0.2741688438 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 8 |
Hb_024621_030 |
0.2797794382 |
- |
- |
tetraketide alpha-pyrone reductase 2 [Jatropha curcas] |
| 9 |
Hb_001114_070 |
0.2842917338 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g58400 [Jatropha curcas] |
| 10 |
Hb_002821_020 |
0.2892914707 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter C family member 4 [Vitis vinifera] |
| 11 |
Hb_006198_150 |
0.2920723013 |
- |
- |
- |
| 12 |
Hb_007919_040 |
0.2939234482 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 13 |
Hb_004195_110 |
0.2943579943 |
- |
- |
hypothetical protein JCGZ_24010 [Jatropha curcas] |
| 14 |
Hb_140049_050 |
0.2944992633 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 15 |
Hb_000928_040 |
0.2953391868 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_005389_190 |
0.2958285142 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 17 |
Hb_002110_260 |
0.2967682585 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_001433_180 |
0.2988853496 |
- |
- |
hypothetical protein JCGZ_23261 [Jatropha curcas] |
| 19 |
Hb_000165_210 |
0.2989087526 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002656_010 |
0.3004707264 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |