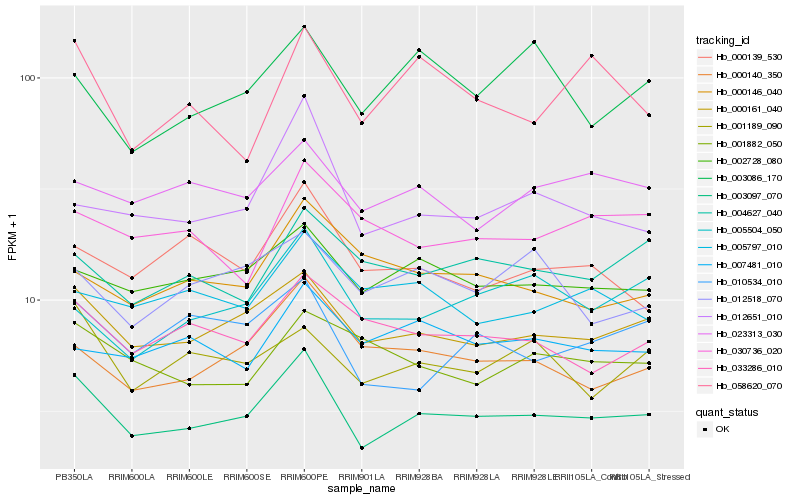
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003097_070 |
0.0 |
- |
- |
glucose regulated repressor protein, putative [Ricinus communis] |
| 2 |
Hb_000161_040 |
0.0733070445 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 3 |
Hb_002728_080 |
0.1048532327 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_012651_010 |
0.1101612182 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 5 |
Hb_000140_350 |
0.1112413224 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010534_010 |
0.1144471582 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 7 |
Hb_033286_010 |
0.115443195 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 8 |
Hb_001189_090 |
0.1160342472 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 9 |
Hb_023313_030 |
0.1184405185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004627_040 |
0.1193127587 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 11 |
Hb_005504_050 |
0.1194034124 |
- |
- |
putative protein [Arabidopsis thaliana] |
| 12 |
Hb_000139_530 |
0.122272085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_058620_070 |
0.1243109879 |
- |
- |
unnamed protein product [Chondrus crispus] |
| 14 |
Hb_001882_050 |
0.1272152533 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_005797_010 |
0.1272287568 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_030736_020 |
0.127786266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_012518_070 |
0.1279020865 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007481_010 |
0.1279603053 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 19 |
Hb_000146_040 |
0.1282103122 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 20 |
Hb_003086_170 |
0.1286325462 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |