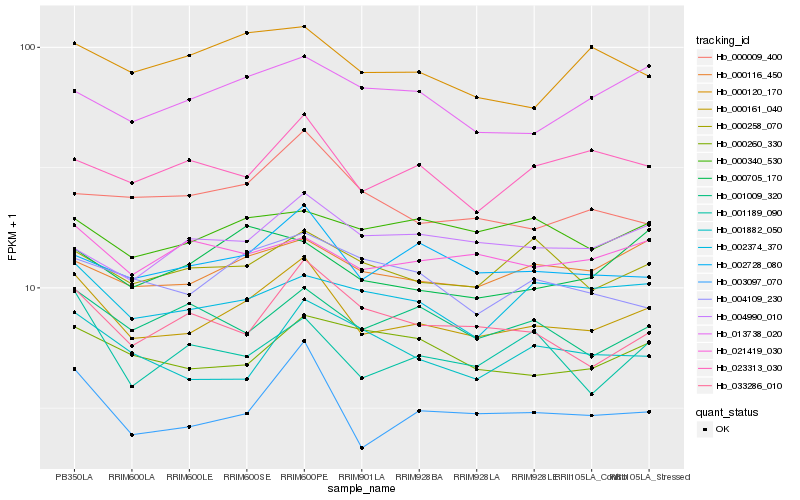
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000161_040 |
0.0 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 2 |
Hb_003097_070 |
0.0733070445 |
- |
- |
glucose regulated repressor protein, putative [Ricinus communis] |
| 3 |
Hb_002728_080 |
0.0769828655 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_000116_450 |
0.0854813712 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_033286_010 |
0.0872718093 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 6 |
Hb_023313_030 |
0.0901900098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000258_070 |
0.0914918913 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 8 |
Hb_002374_370 |
0.0920151332 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_013738_020 |
0.0926708255 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 10 |
Hb_001009_320 |
0.0927408742 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 11 |
Hb_021419_030 |
0.0928463307 |
- |
- |
hypothetical protein glysoja_023295 [Glycine soja] |
| 12 |
Hb_004109_230 |
0.0935883904 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_000009_400 |
0.0936350172 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 14 |
Hb_000260_330 |
0.093744153 |
- |
- |
PREDICTED: uncharacterized protein LOC103438625 [Malus domestica] |
| 15 |
Hb_000120_170 |
0.0943435422 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 16 |
Hb_001189_090 |
0.0945235723 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 17 |
Hb_001882_050 |
0.0963582757 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000705_170 |
0.0963892996 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870 [Jatropha curcas] |
| 19 |
Hb_004990_010 |
0.097384165 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_000340_530 |
0.0978959071 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |