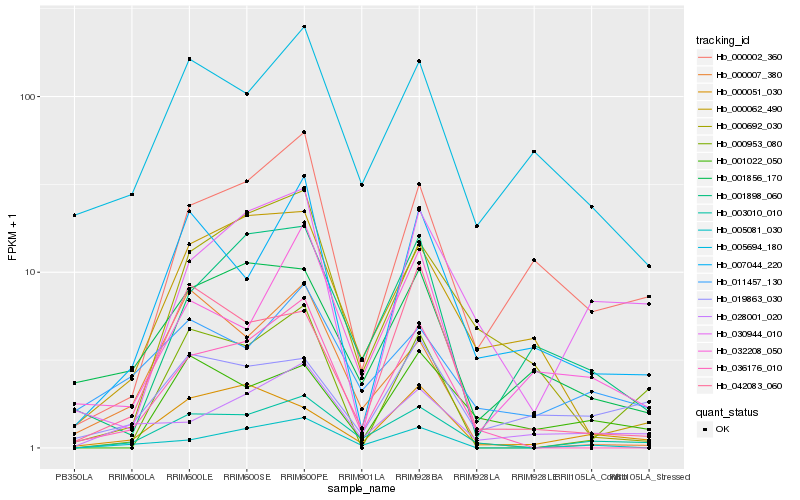
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003010_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633673 [Jatropha curcas] |
| 2 |
Hb_036176_010 |
0.1951360115 |
- |
- |
hypothetical protein CISIN_1g002055mg [Citrus sinensis] |
| 3 |
Hb_011457_130 |
0.2016930176 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 4 |
Hb_005081_030 |
0.2023508187 |
- |
- |
hypothetical protein POPTR_0018s09200g [Populus trichocarpa] |
| 5 |
Hb_007044_220 |
0.2111622297 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_005694_180 |
0.2148351538 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 7 |
Hb_000051_030 |
0.2162174176 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 8 |
Hb_028001_020 |
0.2173640562 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 9 |
Hb_030944_010 |
0.2185654316 |
- |
- |
PREDICTED: uncharacterized protein LOC105633455 [Jatropha curcas] |
| 10 |
Hb_000062_490 |
0.220091085 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000002_360 |
0.2216343078 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 12 |
Hb_001856_170 |
0.2254103254 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 13 |
Hb_032208_050 |
0.2269914181 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 14 |
Hb_000007_380 |
0.2270750494 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 15 |
Hb_000953_080 |
0.2270879902 |
- |
- |
PREDICTED: PAN domain-containing protein At5g03700 [Jatropha curcas] |
| 16 |
Hb_000692_030 |
0.2286147559 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 17 |
Hb_001022_050 |
0.229281174 |
- |
- |
hypothetical protein POPTR_0016s07780g [Populus trichocarpa] |
| 18 |
Hb_019863_030 |
0.2313062157 |
- |
- |
hypothetical protein JCGZ_00393 [Jatropha curcas] |
| 19 |
Hb_001898_060 |
0.2331180388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_042083_060 |
0.2344978707 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |