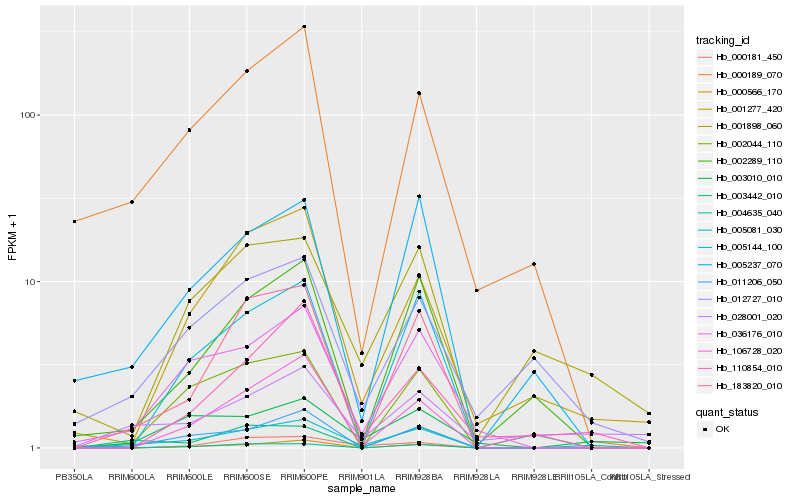
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005081_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s09200g [Populus trichocarpa] |
| 2 |
Hb_036176_010 |
0.1899236346 |
- |
- |
hypothetical protein CISIN_1g002055mg [Citrus sinensis] |
| 3 |
Hb_011206_050 |
0.1908071714 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 4 |
Hb_003010_010 |
0.2023508187 |
- |
- |
PREDICTED: uncharacterized protein LOC105633673 [Jatropha curcas] |
| 5 |
Hb_002044_110 |
0.2099338472 |
- |
- |
PREDICTED: probable carboxylesterase 5 isoform X1 [Populus euphratica] |
| 6 |
Hb_183820_010 |
0.212786989 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 7 |
Hb_003442_010 |
0.2149626168 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 4-like isoform X2 [Citrus sinensis] |
| 8 |
Hb_005237_070 |
0.2164166989 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 9 |
Hb_001277_420 |
0.2205687164 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 10 |
Hb_106728_020 |
0.2222760019 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_028001_020 |
0.2252590084 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 12 |
Hb_005144_100 |
0.2260294026 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_000189_070 |
0.2285265379 |
transcription factor |
TF Family: AUX/IAA |
Auxin-induced protein 22D, putative [Ricinus communis] |
| 14 |
Hb_002289_110 |
0.2287630715 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 15 |
Hb_000566_170 |
0.2292611318 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 16 |
Hb_001898_060 |
0.2297599519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004635_040 |
0.2306506995 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 18 |
Hb_012727_010 |
0.2307155947 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 19 |
Hb_000181_450 |
0.2316581802 |
- |
- |
hypothetical protein JCGZ_20984 [Jatropha curcas] |
| 20 |
Hb_110854_010 |
0.2322571414 |
- |
- |
- |