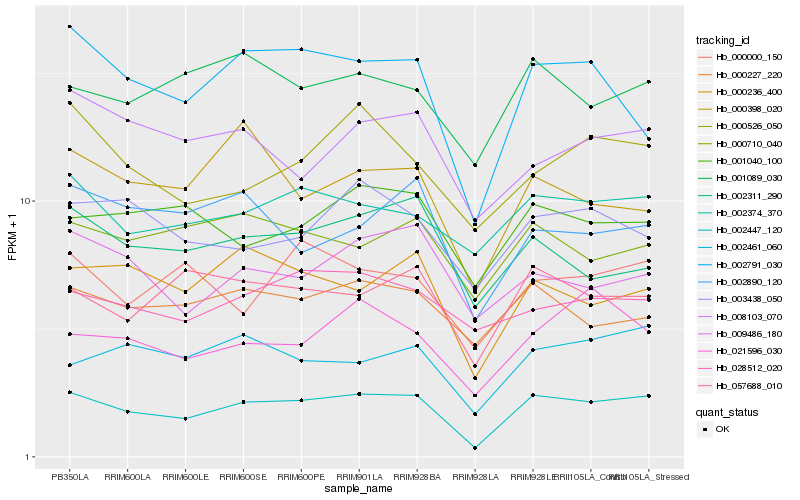
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002447_120 |
0.0 |
- |
- |
hypothetical protein O988_01801, partial [Pseudogymnoascus pannorum VKM F-3808] |
| 2 |
Hb_002791_030 |
0.1093897808 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 3 |
Hb_000236_400 |
0.1136113247 |
- |
- |
hypothetical protein JCGZ_01058 [Jatropha curcas] |
| 4 |
Hb_002461_060 |
0.1179088506 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 5 |
Hb_057688_010 |
0.1180243444 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002374_370 |
0.1189241669 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_021596_030 |
0.1241014376 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001089_030 |
0.12435159 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 9 |
Hb_000227_220 |
0.1260736856 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 10 |
Hb_001040_100 |
0.1270957755 |
- |
- |
PREDICTED: general transcription factor IIH subunit 4 [Jatropha curcas] |
| 11 |
Hb_002311_290 |
0.1277638181 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 12 |
Hb_009486_180 |
0.128215083 |
- |
- |
unknown [Glycine max] |
| 13 |
Hb_000526_050 |
0.1284873648 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_003438_050 |
0.1286416644 |
- |
- |
PREDICTED: rab3 GTPase-activating protein catalytic subunit isoform X2 [Jatropha curcas] |
| 15 |
Hb_000710_040 |
0.1286587429 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 16 |
Hb_000000_150 |
0.128909625 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 17 |
Hb_002890_120 |
0.1294752634 |
- |
- |
Nucleic acid binding,RNA binding isoform 1 [Theobroma cacao] |
| 18 |
Hb_000398_020 |
0.1303621829 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008103_070 |
0.1317099562 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 20 |
Hb_028512_020 |
0.1321880358 |
- |
- |
PREDICTED: protein S-acyltransferase 11 isoform X2 [Jatropha curcas] |