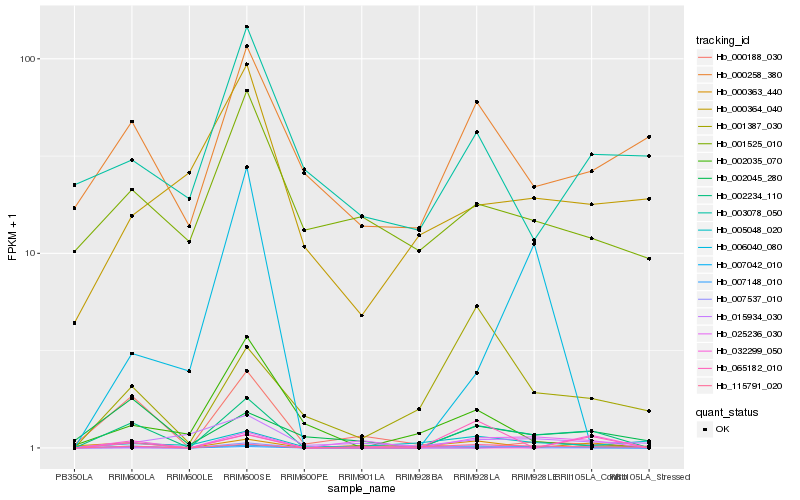
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002234_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103455142 [Malus domestica] |
| 2 |
Hb_002045_280 |
0.3677626006 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_115791_020 |
0.3788676841 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 4 |
Hb_007042_010 |
0.3912921605 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_000363_440 |
0.3957754395 |
- |
- |
- |
| 6 |
Hb_065182_010 |
0.3961564179 |
- |
- |
hypothetical protein JCGZ_20878 [Jatropha curcas] |
| 7 |
Hb_001387_030 |
0.3986192539 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0013s13920g [Populus trichocarpa] |
| 8 |
Hb_007148_010 |
0.4185279873 |
- |
- |
PREDICTED: uncharacterized protein LOC103427080 [Malus domestica] |
| 9 |
Hb_000258_380 |
0.4203934683 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP11-like [Jatropha curcas] |
| 10 |
Hb_032299_050 |
0.4217448593 |
- |
- |
- |
| 11 |
Hb_006040_080 |
0.4271270051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002035_070 |
0.4277536692 |
- |
- |
hypothetical protein CICLE_v10033569mg [Citrus clementina] |
| 13 |
Hb_025236_030 |
0.4358462153 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 14 |
Hb_015934_030 |
0.4363337886 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_000188_030 |
0.4394088359 |
- |
- |
carboxylic ester hydrolase, putative [Ricinus communis] |
| 16 |
Hb_005048_020 |
0.4402515734 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 17 |
Hb_007537_010 |
0.4405212404 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_001525_010 |
0.4418703273 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 19 |
Hb_000364_040 |
0.4434710717 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF010-like [Jatropha curcas] |
| 20 |
Hb_003078_050 |
0.4466414628 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |