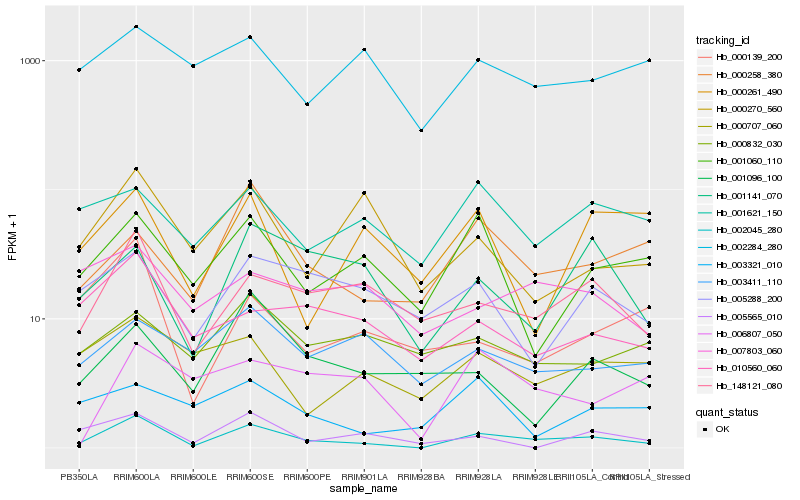
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002045_280 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_148121_080 |
0.2419236157 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SHOOT GRAVITROPISM 5 [Populus euphratica] |
| 3 |
Hb_000258_380 |
0.2723354917 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP11-like [Jatropha curcas] |
| 4 |
Hb_001060_110 |
0.2788190197 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 5 |
Hb_001141_070 |
0.2791749101 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 3 [Jatropha curcas] |
| 6 |
Hb_010560_060 |
0.2811468368 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 7 |
Hb_000139_200 |
0.2840580842 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_005565_010 |
0.2882407515 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 9 |
Hb_005288_200 |
0.2885902763 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 10 |
Hb_001621_150 |
0.29137465 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Jatropha curcas] |
| 11 |
Hb_000270_560 |
0.2929134341 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |
| 12 |
Hb_000707_060 |
0.29484003 |
- |
- |
- |
| 13 |
Hb_003411_110 |
0.2965258881 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 14 |
Hb_001096_100 |
0.2987144889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007803_060 |
0.299385615 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 16 |
Hb_006807_050 |
0.3002353594 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPH8A, putative [Ricinus communis] |
| 17 |
Hb_000261_490 |
0.3040131521 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 18 |
Hb_000832_030 |
0.3049608062 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634915 [Jatropha curcas] |
| 19 |
Hb_002284_280 |
0.3050334269 |
transcription factor |
TF Family: HMG |
high mobility group protein [Hevea brasiliensis] |
| 20 |
Hb_003321_010 |
0.3058335788 |
- |
- |
hypothetical protein RCOM_0884420 [Ricinus communis] |