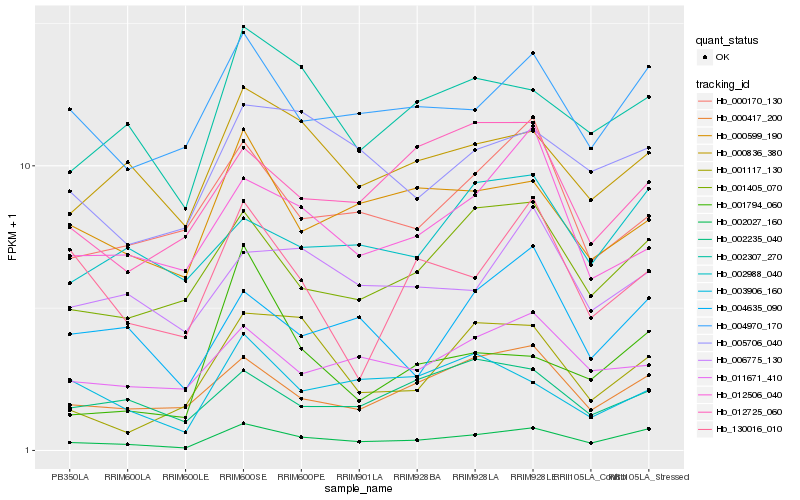
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002027_160 |
0.0 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_000417_200 |
0.1364980737 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001405_070 |
0.1380401929 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 4 |
Hb_004970_170 |
0.1413249017 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 5 |
Hb_002307_270 |
0.1461289491 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 6 |
Hb_004635_090 |
0.1552694094 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 7 |
Hb_000836_380 |
0.156430204 |
- |
- |
PREDICTED: C-terminal binding protein AN-like [Populus euphratica] |
| 8 |
Hb_011671_410 |
0.1600867291 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 9 |
Hb_005706_040 |
0.1611255182 |
- |
- |
PREDICTED: uncharacterized protein LOC105642418 [Jatropha curcas] |
| 10 |
Hb_000599_190 |
0.1614864449 |
transcription factor |
TF Family: SNF2 |
DNA repair and recombination protein RAD26, putative [Ricinus communis] |
| 11 |
Hb_002988_040 |
0.164388791 |
- |
- |
- |
| 12 |
Hb_000170_130 |
0.1649693699 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 13 |
Hb_001117_130 |
0.1650980722 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 14 |
Hb_002235_040 |
0.1667892077 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 15 |
Hb_006775_130 |
0.1679280982 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 16 |
Hb_001794_060 |
0.1700874069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_012725_060 |
0.171629171 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_012506_040 |
0.1718253665 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 19 |
Hb_130016_010 |
0.171896949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003906_160 |
0.1725592037 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |