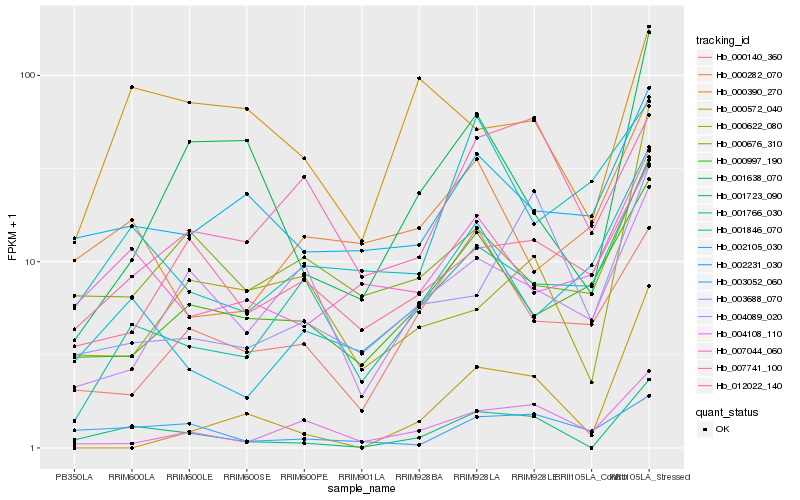
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001723_090 |
0.0 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003052_060 |
0.2684881753 |
- |
- |
PREDICTED: uncharacterized protein LOC105640717 [Jatropha curcas] |
| 3 |
Hb_001638_070 |
0.2875264112 |
transcription factor |
TF Family: bHLH |
Basic helix-loop-helix DNA-binding family protein [Theobroma cacao] |
| 4 |
Hb_002231_030 |
0.2880024235 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 5 |
Hb_003688_070 |
0.2884165863 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein E [Jatropha curcas] |
| 6 |
Hb_004108_110 |
0.2900132924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_012022_140 |
0.2928198279 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 8 |
Hb_001766_030 |
0.2983791305 |
- |
- |
hypothetical protein POPTR_0017s12640g [Populus trichocarpa] |
| 9 |
Hb_007044_060 |
0.3000709806 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_000997_190 |
0.30462823 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like [Jatropha curcas] |
| 11 |
Hb_002105_030 |
0.3081239125 |
- |
- |
- |
| 12 |
Hb_000572_040 |
0.3083770829 |
- |
- |
hypothetical protein POPTR_0006s14815g [Populus trichocarpa] |
| 13 |
Hb_000622_080 |
0.3106147075 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 14 |
Hb_000390_270 |
0.3115681827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000140_360 |
0.3132430486 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 16 |
Hb_004089_020 |
0.3140626869 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 17 |
Hb_000676_310 |
0.3174977011 |
- |
- |
BnaC09g40680D [Brassica napus] |
| 18 |
Hb_007741_100 |
0.3199453335 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 19 |
Hb_001846_070 |
0.3208543905 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 20 |
Hb_000282_070 |
0.3217395757 |
- |
- |
PREDICTED: protein REVERSION-TO-ETHYLENE SENSITIVITY1 [Jatropha curcas] |