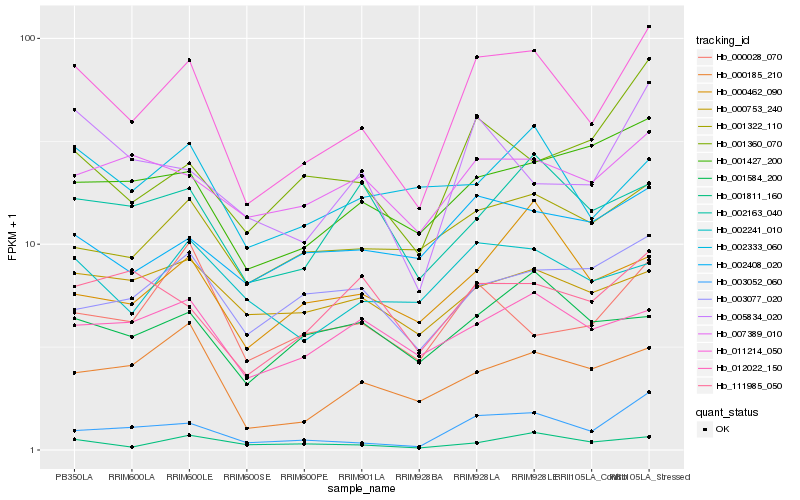
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011214_050 |
0.0 |
- |
- |
phytoene synthase 2 [Manihot esculenta] |
| 2 |
Hb_003052_060 |
0.1396997735 |
- |
- |
PREDICTED: uncharacterized protein LOC105640717 [Jatropha curcas] |
| 3 |
Hb_001427_200 |
0.1503483324 |
- |
- |
PREDICTED: uncharacterized protein LOC105637796 [Jatropha curcas] |
| 4 |
Hb_005834_020 |
0.1510007397 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001322_110 |
0.1515479734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001811_160 |
0.1579295573 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Eucalyptus grandis] |
| 7 |
Hb_000185_210 |
0.1580195684 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002241_010 |
0.1583650345 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 9 |
Hb_001360_070 |
0.1585075252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012022_150 |
0.1591599285 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |
| 11 |
Hb_002408_020 |
0.1594022129 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 12 |
Hb_002163_040 |
0.1607817837 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007389_010 |
0.1613800259 |
- |
- |
PREDICTED: THO complex subunit 7A [Jatropha curcas] |
| 14 |
Hb_111985_050 |
0.1643335914 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_002333_060 |
0.1643372274 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_003077_020 |
0.1646814791 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_000753_240 |
0.1663246689 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 18 |
Hb_000462_090 |
0.1669724861 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001584_200 |
0.1669793034 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 20 |
Hb_000028_070 |
0.1687082106 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |