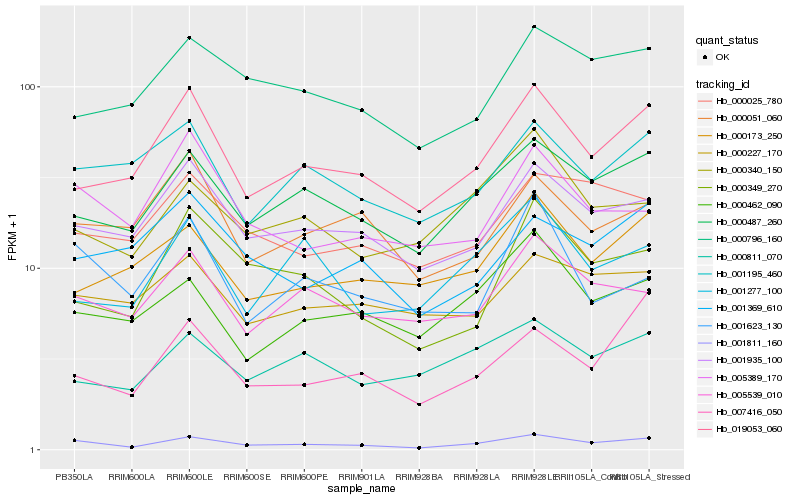
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001811_160 |
0.0 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Eucalyptus grandis] |
| 2 |
Hb_000487_260 |
0.1123624009 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001935_100 |
0.1237540458 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 4 |
Hb_019053_060 |
0.1265751483 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 5 |
Hb_005539_010 |
0.1429015598 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 6 |
Hb_000462_090 |
0.1447385327 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001195_460 |
0.1452439478 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000796_160 |
0.146836345 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000025_780 |
0.1484331967 |
- |
- |
PREDICTED: sanguinarine reductase [Jatropha curcas] |
| 10 |
Hb_000349_270 |
0.1492918 |
- |
- |
PREDICTED: uncharacterized protein ycf49 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000340_150 |
0.1493623789 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 12 |
Hb_005389_170 |
0.1499094457 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 13 |
Hb_000811_070 |
0.1504261909 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 14 |
Hb_000227_170 |
0.1532426252 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_007416_050 |
0.153678565 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 5 [Jatropha curcas] |
| 16 |
Hb_001623_130 |
0.1550494822 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000051_060 |
0.155640569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001277_100 |
0.156788191 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 19 |
Hb_001369_610 |
0.1575565295 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000173_250 |
0.1576013104 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |