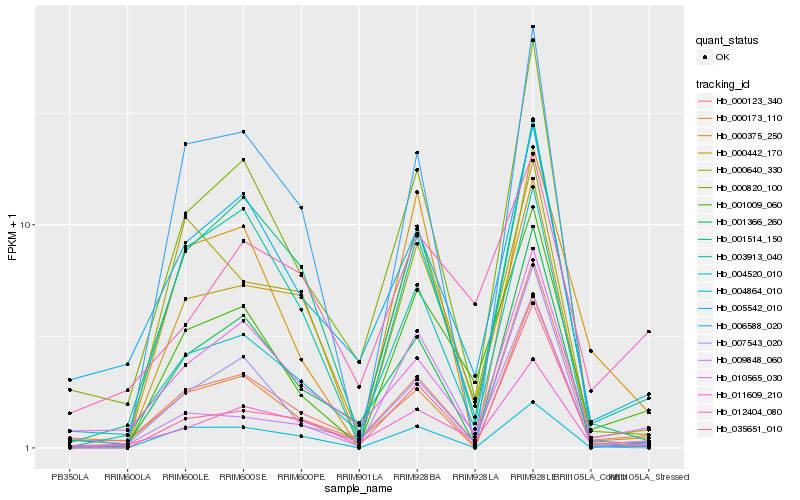
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001009_060 |
0.0 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 [Jatropha curcas] |
| 2 |
Hb_003913_040 |
0.1369502843 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 3 |
Hb_011609_210 |
0.1773389047 |
- |
- |
Uncharacterized protein TCM_011393 [Theobroma cacao] |
| 4 |
Hb_000820_100 |
0.1818936977 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 5 |
Hb_000442_170 |
0.182194392 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 6 |
Hb_000123_340 |
0.1833147677 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 7 |
Hb_001514_150 |
0.1901708482 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 8 |
Hb_000375_250 |
0.1903214049 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 9 |
Hb_004520_010 |
0.1910791998 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 10 |
Hb_010565_030 |
0.193219796 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 11 |
Hb_035651_010 |
0.1968409459 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 12 |
Hb_005542_010 |
0.1972097036 |
- |
- |
Uncharacterized protein TCM_021447 [Theobroma cacao] |
| 13 |
Hb_001366_260 |
0.2025025808 |
- |
- |
T4.15 [Malus x robusta] |
| 14 |
Hb_000640_330 |
0.2034180034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000173_110 |
0.2084663862 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 16 |
Hb_007543_020 |
0.2097330555 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 17 |
Hb_004864_010 |
0.2117577326 |
- |
- |
hypothetical protein CICLE_v10003960mg, partial [Citrus clementina] |
| 18 |
Hb_012404_080 |
0.2120387689 |
- |
- |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 19 |
Hb_006588_020 |
0.2134973181 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_009848_060 |
0.2142098995 |
- |
- |
unknown [Glycine max] |