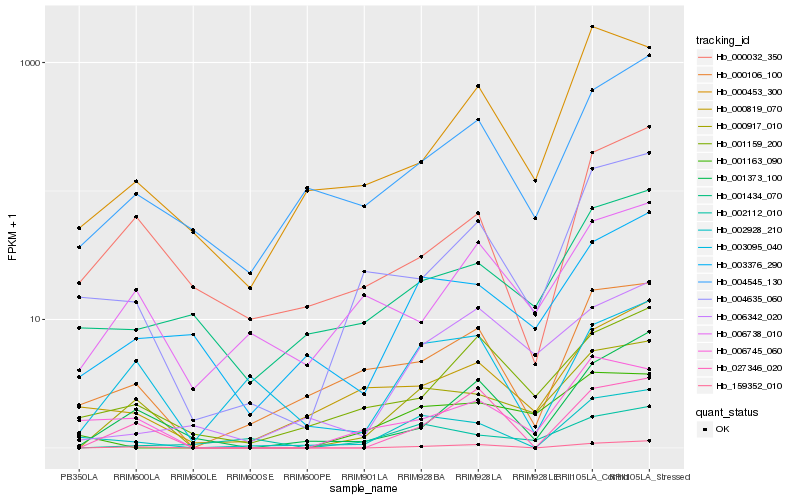
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000917_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_159352_010 |
0.2219730459 |
- |
- |
hypothetical protein JCGZ_12393 [Jatropha curcas] |
| 3 |
Hb_006745_060 |
0.2268311081 |
- |
- |
- |
| 4 |
Hb_003376_290 |
0.228580234 |
- |
- |
hypothetical protein POPTR_0010s22880g [Populus trichocarpa] |
| 5 |
Hb_004635_060 |
0.2399571769 |
- |
- |
hypothetical protein POPTR_0006s22850g [Populus trichocarpa] |
| 6 |
Hb_001373_100 |
0.241007411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001159_200 |
0.253050923 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_004545_130 |
0.2566275178 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 9 |
Hb_027346_020 |
0.2572163114 |
- |
- |
- |
| 10 |
Hb_000106_100 |
0.2577230149 |
- |
- |
- |
| 11 |
Hb_001163_090 |
0.2600368614 |
- |
- |
hypothetical protein POPTR_0009s14920g [Populus trichocarpa] |
| 12 |
Hb_001434_070 |
0.260419076 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 13 |
Hb_006342_020 |
0.2606054391 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000819_070 |
0.2620108421 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 15 |
Hb_006738_010 |
0.262520052 |
- |
- |
PREDICTED: uncharacterized protein LOC105634993 [Jatropha curcas] |
| 16 |
Hb_003095_040 |
0.264222566 |
- |
- |
hypothetical protein CICLE_v10030016mg [Citrus clementina] |
| 17 |
Hb_002112_010 |
0.2652069666 |
- |
- |
hypothetical protein POPTR_0001s45830g [Populus trichocarpa] |
| 18 |
Hb_000453_300 |
0.2652088387 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 19 |
Hb_002928_210 |
0.266900821 |
- |
- |
- |
| 20 |
Hb_000032_350 |
0.268367335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |