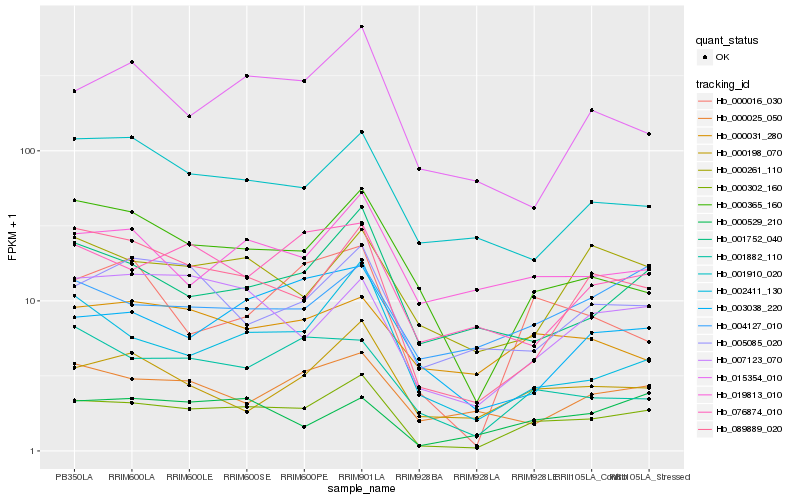
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648403 [Jatropha curcas] |
| 2 |
Hb_089889_020 |
0.1512008725 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At2g36080-like [Jatropha curcas] |
| 3 |
Hb_001752_040 |
0.1609263949 |
- |
- |
hypothetical protein EUGRSUZ_C02580 [Eucalyptus grandis] |
| 4 |
Hb_000365_160 |
0.1613158055 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 5 |
Hb_007123_070 |
0.1621231528 |
- |
- |
- |
| 6 |
Hb_002411_130 |
0.1658063694 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Populus euphratica] |
| 7 |
Hb_000198_070 |
0.1685548594 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 8 |
Hb_019813_010 |
0.1714126859 |
- |
- |
PREDICTED: uncharacterized protein LOC105646947 [Jatropha curcas] |
| 9 |
Hb_005085_020 |
0.1735573355 |
- |
- |
hypothetical protein B456_001G193200 [Gossypium raimondii] |
| 10 |
Hb_015354_010 |
0.1744892852 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 11 |
Hb_000016_030 |
0.1745071618 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_076874_010 |
0.1754167602 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 13 |
Hb_003038_220 |
0.1755245264 |
- |
- |
eukaryotic translation initiation factor 2 gamma subunit, putative [Ricinus communis] |
| 14 |
Hb_000529_210 |
0.1766213045 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 15 |
Hb_000261_110 |
0.1770616845 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 16 |
Hb_000025_050 |
0.1798178103 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 17 |
Hb_001882_110 |
0.1798830373 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004127_010 |
0.1804771075 |
- |
- |
PREDICTED: probable RNA 3'-terminal phosphate cyclase-like protein [Malus domestica] |
| 19 |
Hb_001910_020 |
0.1810702199 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 20 |
Hb_000031_280 |
0.1815918457 |
- |
- |
- |