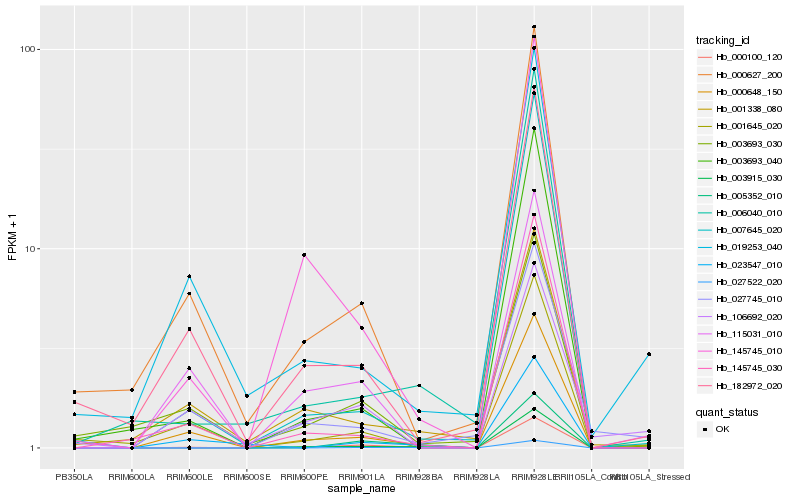
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000100_120 |
0.0 |
- |
- |
RNA-directed DNA polymerase (Reverse transcriptase); Chromo; Zinc finger, CCHC-type; Peptidase aspartic, active site; Polynucleotidyl transferase, Ribonuclease H fold [Medicago truncatula] |
| 2 |
Hb_000648_150 |
0.1495346994 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase V.1 [Camelina sativa] |
| 3 |
Hb_003915_030 |
0.1596164747 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_005352_010 |
0.1669781365 |
- |
- |
- |
| 5 |
Hb_115031_010 |
0.1683700951 |
- |
- |
photosystem II P680 chlorophyll A apoprotein, partial (chloroplast) [Oenothera grandiflora] |
| 6 |
Hb_027522_020 |
0.1720117289 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_000627_200 |
0.1790216144 |
- |
- |
NADH dehydrogenase subunit I [Hevea brasiliensis] |
| 8 |
Hb_001645_020 |
0.1813298282 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit H [Medicago truncatula] |
| 9 |
Hb_182972_020 |
0.1842473505 |
- |
- |
cytochrome b6 [Gossypium barbadense] |
| 10 |
Hb_145745_010 |
0.1847672916 |
- |
- |
BnaUnng00830D [Brassica napus] |
| 11 |
Hb_145745_030 |
0.1866181998 |
- |
- |
BnaC09g26890D [Brassica napus] |
| 12 |
Hb_106692_020 |
0.1868153447 |
- |
- |
NADH dehydrogenase subunit F, partial (chloroplast) [Hevea sp. Gillespie 4272] |
| 13 |
Hb_007645_020 |
0.1868432566 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit [Medicago truncatula] |
| 14 |
Hb_019253_040 |
0.1884988374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006040_010 |
0.1885624809 |
- |
- |
cytochrome c oxidase subunit 3 (mitochondrion) [Hevea brasiliensis] |
| 16 |
Hb_003693_030 |
0.1894538483 |
- |
- |
NADH dehydrogenase subunit 7 [Hevea brasiliensis] |
| 17 |
Hb_003693_040 |
0.1916414337 |
- |
- |
NADH dehydrogenase subunit 1 [Hevea brasiliensis] |
| 18 |
Hb_027745_010 |
0.1935299995 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 19 |
Hb_023547_010 |
0.1951773026 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_001338_080 |
0.1969075946 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Tamarindus indica] |