| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
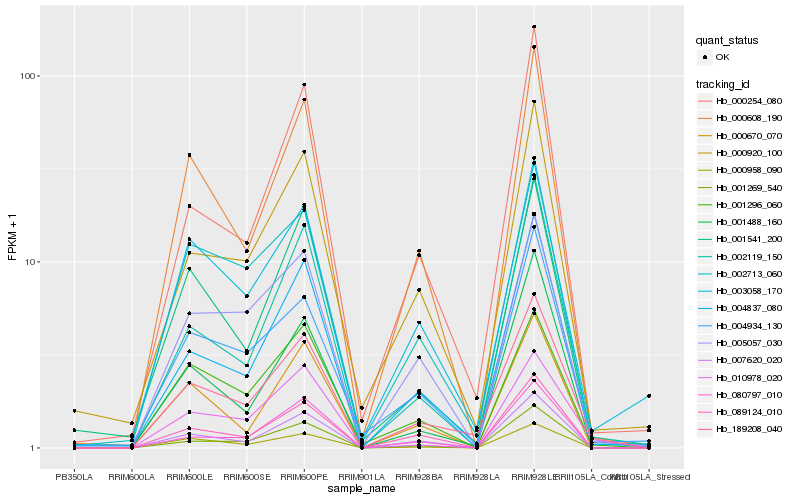
Hb_189208_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |
| 2 |
Hb_004837_080 |
0.1124898618 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 3 |
Hb_000608_190 |
0.1231846725 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 4 |
Hb_007620_020 |
0.1267607282 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000920_100 |
0.1291000181 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_089124_010 |
0.1301002663 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 7 |
Hb_002713_060 |
0.1330213084 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 8 |
Hb_000254_080 |
0.1343143733 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 9 |
Hb_001269_540 |
0.134517401 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 10 |
Hb_002119_150 |
0.1358636955 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 11 |
Hb_003058_170 |
0.13829803 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005057_030 |
0.1406528523 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 13 |
Hb_010978_020 |
0.1418531652 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL12 [Jatropha curcas] |
| 14 |
Hb_000958_090 |
0.1418848958 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 15 |
Hb_004934_130 |
0.1437396025 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 16 |
Hb_001541_200 |
0.1437889868 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 17 |
Hb_001296_060 |
0.1464920804 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 18 |
Hb_000670_070 |
0.1474323292 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 19 |
Hb_001488_160 |
0.1477077807 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_080797_010 |
0.1478411912 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |