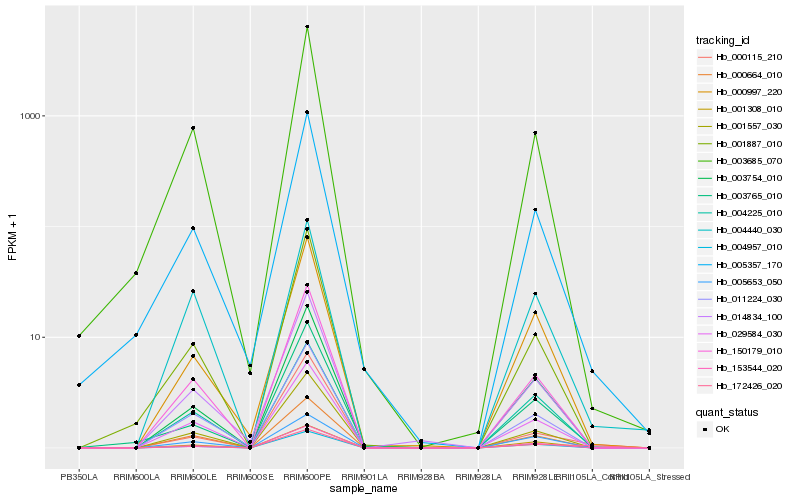
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172426_020 |
0.0 |
- |
- |
PREDICTED: putative fasciclin-like arabinogalactan protein 20 [Jatropha curcas] |
| 2 |
Hb_000115_210 |
0.196087697 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 3-like [Jatropha curcas] |
| 3 |
Hb_005357_170 |
0.196166457 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 4 |
Hb_150179_010 |
0.1971147116 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |
| 5 |
Hb_004957_010 |
0.1971725178 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 6 |
Hb_000997_220 |
0.1978505473 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 7 |
Hb_029584_030 |
0.1979569554 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 8 |
Hb_003754_010 |
0.1983037799 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 9 |
Hb_001308_010 |
0.199763752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000664_010 |
0.2008299774 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |
| 11 |
Hb_004440_030 |
0.2011706651 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 12 |
Hb_011224_030 |
0.2017390707 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001557_030 |
0.2026210778 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 14 |
Hb_153544_020 |
0.2056833222 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP12 [Jatropha curcas] |
| 15 |
Hb_004225_010 |
0.2060166739 |
- |
- |
- |
| 16 |
Hb_014834_100 |
0.2074435232 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 17 |
Hb_003685_070 |
0.2075287841 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 18 |
Hb_005653_050 |
0.2083364488 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3-sialyltransferase [Jatropha curcas] |
| 19 |
Hb_003765_010 |
0.2097811514 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 20 |
Hb_001887_010 |
0.2106114038 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |