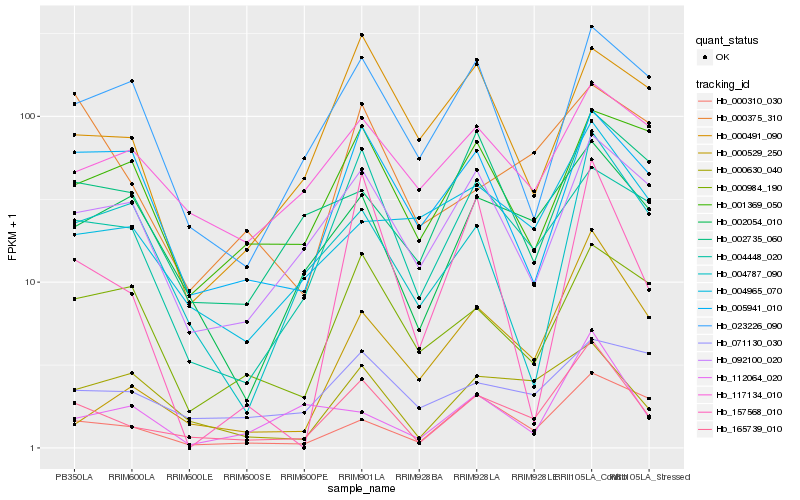
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_165739_010 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000630_040 |
0.2089833146 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 3 |
Hb_002054_010 |
0.20982076 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 4 |
Hb_000310_030 |
0.2139774288 |
- |
- |
hypothetical protein JCGZ_03883 [Jatropha curcas] |
| 5 |
Hb_092100_020 |
0.2192096031 |
- |
- |
strictosidine synthase, putative [Ricinus communis] |
| 6 |
Hb_004448_020 |
0.2249175484 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000529_250 |
0.2317538204 |
- |
- |
PREDICTED: nicotinamidase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_005941_010 |
0.234607526 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 9 |
Hb_004965_070 |
0.2386546326 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 10 |
Hb_023226_090 |
0.2387310811 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 11 |
Hb_000984_190 |
0.2389658534 |
- |
- |
- |
| 12 |
Hb_157568_010 |
0.2413506124 |
- |
- |
MATE efflux family protein, putative [Theobroma cacao] |
| 13 |
Hb_117134_010 |
0.241822887 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein isoform X1 [Jatropha curcas] |
| 14 |
Hb_004787_090 |
0.2434919482 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 15 |
Hb_002735_060 |
0.2463317617 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 16 |
Hb_000375_310 |
0.2470430839 |
- |
- |
hypothetical protein JCGZ_18643 [Jatropha curcas] |
| 17 |
Hb_071130_030 |
0.2487078992 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 18 |
Hb_112064_020 |
0.2487863027 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 19 |
Hb_001369_050 |
0.250613719 |
- |
- |
hypothetical protein MIMGU_mgv1a0083172mg, partial [Erythranthe guttata] |
| 20 |
Hb_000491_090 |
0.2519670973 |
- |
- |
hypothetical protein B456_008G219400 [Gossypium raimondii] |