| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
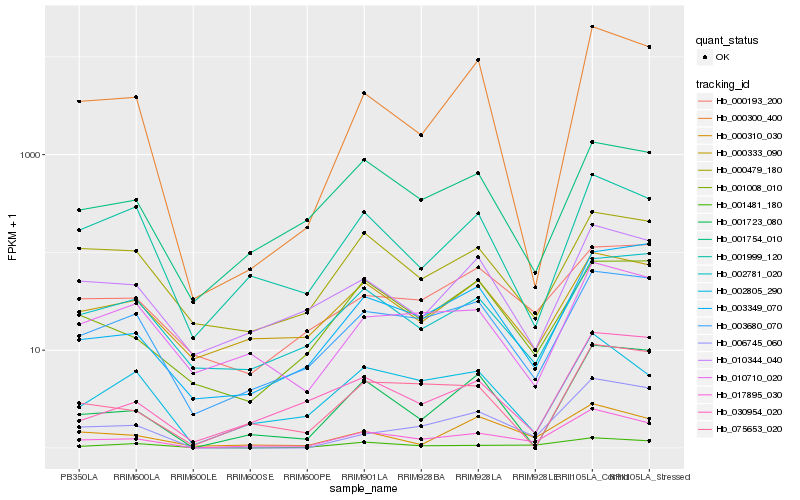
Hb_017895_030 |
0.0 |
transcription factor |
TF Family: C2H2 |
- |
| 2 |
Hb_003680_070 |
0.1437356542 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001754_010 |
0.1673882525 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 4 |
Hb_001008_010 |
0.1714179839 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic-like [Populus euphratica] |
| 5 |
Hb_010344_040 |
0.1714788763 |
- |
- |
PREDICTED: uncharacterized protein LOC105636452 [Jatropha curcas] |
| 6 |
Hb_000333_090 |
0.1716301214 |
- |
- |
PREDICTED: uncharacterized protein LOC102617374 isoform X1 [Citrus sinensis] |
| 7 |
Hb_000310_030 |
0.172446777 |
- |
- |
hypothetical protein JCGZ_03883 [Jatropha curcas] |
| 8 |
Hb_001481_180 |
0.1730811207 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 9 |
Hb_003349_070 |
0.1748896909 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 10 |
Hb_010710_020 |
0.1780693944 |
- |
- |
PREDICTED: uncharacterized protein LOC103322096 [Prunus mume] |
| 11 |
Hb_030954_020 |
0.1791529844 |
- |
- |
hypothetical protein RCOM_1330200 [Ricinus communis] |
| 12 |
Hb_002781_020 |
0.1796451684 |
- |
- |
PREDICTED: uncharacterized protein LOC105644398 isoform X2 [Jatropha curcas] |
| 13 |
Hb_075653_020 |
0.1804292918 |
- |
- |
- |
| 14 |
Hb_000479_180 |
0.1819586876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006745_060 |
0.1822739909 |
- |
- |
- |
| 16 |
Hb_000193_200 |
0.1823327525 |
- |
- |
rotamase, putative [Ricinus communis] |
| 17 |
Hb_002805_290 |
0.1826776177 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_001723_080 |
0.1830447011 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 19 |
Hb_001999_120 |
0.1856644139 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_000300_400 |
0.1863000816 |
- |
- |
hypothetical protein PHAVU_001G114100g [Phaseolus vulgaris] |