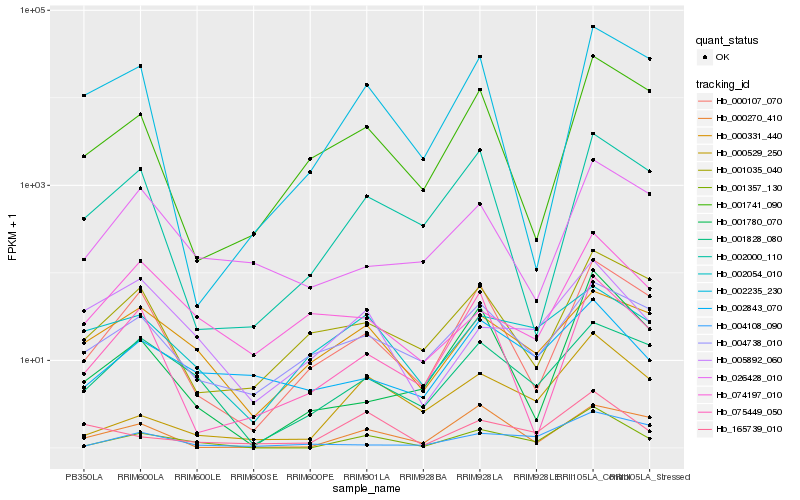
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_00933 [Jatropha curcas] |
| 2 |
Hb_000529_250 |
0.2139908801 |
- |
- |
PREDICTED: nicotinamidase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000107_070 |
0.2271615141 |
- |
- |
PREDICTED: uncharacterized protein LOC105641007 [Jatropha curcas] |
| 4 |
Hb_001828_080 |
0.2354162075 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase family protein [Populus trichocarpa] |
| 5 |
Hb_002843_070 |
0.2417494331 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_001780_070 |
0.2469450373 |
- |
- |
hypothetical protein JCGZ_08549 [Jatropha curcas] |
| 7 |
Hb_075449_050 |
0.2589217406 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 8 |
Hb_004738_010 |
0.2598685024 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 9 |
Hb_002000_110 |
0.2609105955 |
- |
- |
ASR-like protein 2 [Hevea brasiliensis] |
| 10 |
Hb_001741_090 |
0.2619939656 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 11 |
Hb_001035_040 |
0.2667266944 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000331_440 |
0.2682105868 |
- |
- |
hypothetical protein POPTR_0015s10290g [Populus trichocarpa] |
| 13 |
Hb_165739_010 |
0.269366182 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_005892_060 |
0.2699189959 |
- |
- |
PREDICTED: uncharacterized protein LOC105632176 isoform X2 [Jatropha curcas] |
| 15 |
Hb_026428_010 |
0.2717886154 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 16 |
Hb_002235_230 |
0.2737300987 |
- |
- |
latex abundant protein 1 [Hevea brasiliensis] |
| 17 |
Hb_004108_090 |
0.2739598465 |
- |
- |
PREDICTED: uncharacterized protein LOC105650231 [Jatropha curcas] |
| 18 |
Hb_074197_010 |
0.2746534079 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 19 |
Hb_000270_410 |
0.2749420556 |
- |
- |
- |
| 20 |
Hb_002054_010 |
0.2749536493 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |