| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
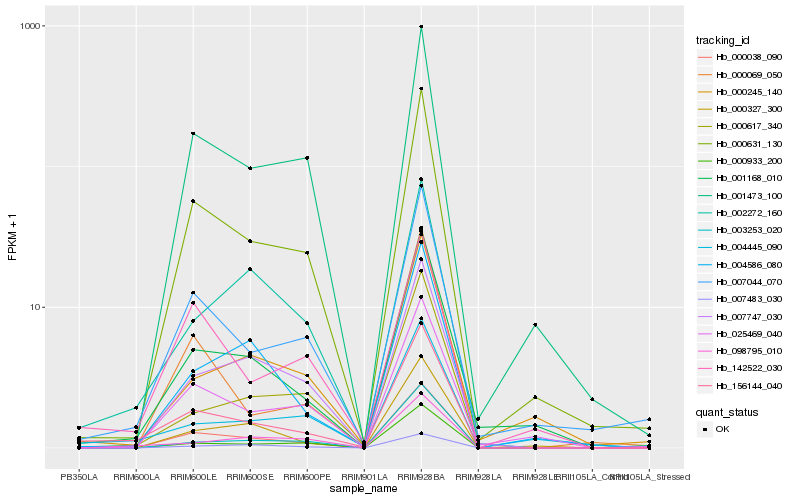
Hb_156144_040 |
0.0 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000038_090 |
0.1272329084 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0002s05820g [Populus trichocarpa] |
| 3 |
Hb_000631_130 |
0.1311505804 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 4 |
Hb_001168_010 |
0.1337512964 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 5 |
Hb_007044_070 |
0.1394051439 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Jatropha curcas] |
| 6 |
Hb_004445_090 |
0.1526508619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000933_200 |
0.1540052025 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_003253_020 |
0.1600592631 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_142522_030 |
0.1636520153 |
- |
- |
hypothetical protein JCGZ_21596 [Jatropha curcas] |
| 10 |
Hb_000327_300 |
0.1655617746 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 11 |
Hb_002272_160 |
0.1660401331 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_004586_080 |
0.1661118312 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 13 |
Hb_000617_340 |
0.1663222164 |
- |
- |
respiratory burst oxidase B [Manihot esculenta] |
| 14 |
Hb_000069_050 |
0.1673758681 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 15 |
Hb_098795_010 |
0.1686932367 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase-like [Jatropha curcas] |
| 16 |
Hb_001473_100 |
0.1696779328 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 17 |
Hb_025469_040 |
0.1702002273 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 18 |
Hb_000245_140 |
0.1724371313 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 19 |
Hb_007747_030 |
0.173431652 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 20 |
Hb_007483_030 |
0.1775810981 |
- |
- |
- |