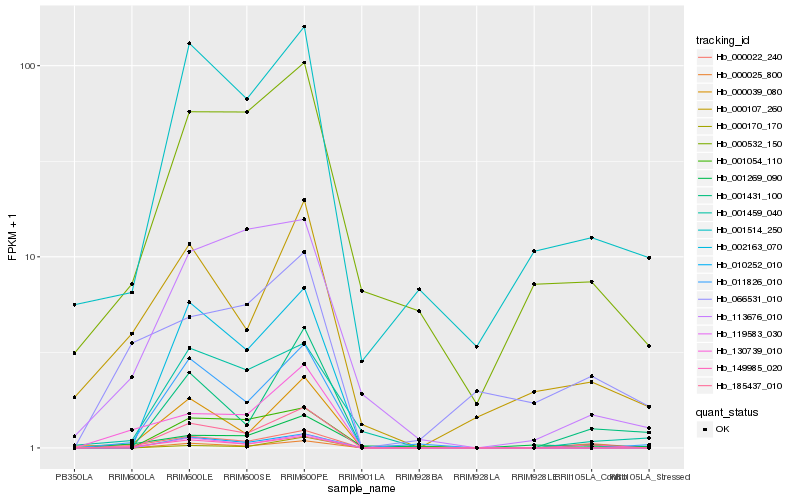
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_149985_020 |
0.0 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Nicotiana tomentosiformis] |
| 2 |
Hb_000022_240 |
0.0909213612 |
- |
- |
PREDICTED: uncharacterized protein LOC105638288 [Jatropha curcas] |
| 3 |
Hb_000107_260 |
0.2546893189 |
- |
- |
- |
| 4 |
Hb_001514_250 |
0.2601988694 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 5 |
Hb_001431_100 |
0.267475892 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 [Jatropha curcas] |
| 6 |
Hb_000170_170 |
0.2677973192 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 7 |
Hb_001054_110 |
0.2707270374 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 8 |
Hb_011826_010 |
0.2714297736 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 9 |
Hb_000532_150 |
0.2754581594 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 10 |
Hb_130739_010 |
0.278028097 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_001459_040 |
0.2791715051 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 12 |
Hb_000039_080 |
0.2829599991 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 13 |
Hb_066531_010 |
0.285399285 |
- |
- |
ccd1, putative [Ricinus communis] |
| 14 |
Hb_010252_010 |
0.2869956266 |
- |
- |
- |
| 15 |
Hb_000025_800 |
0.2870423143 |
- |
- |
hypothetical protein VITISV_020143 [Vitis vinifera] |
| 16 |
Hb_119583_030 |
0.2873987064 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_001269_090 |
0.2879313504 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 18 |
Hb_185437_010 |
0.2880869193 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 19 |
Hb_113676_010 |
0.2884148804 |
- |
- |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 20 |
Hb_002163_070 |
0.2886374395 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |