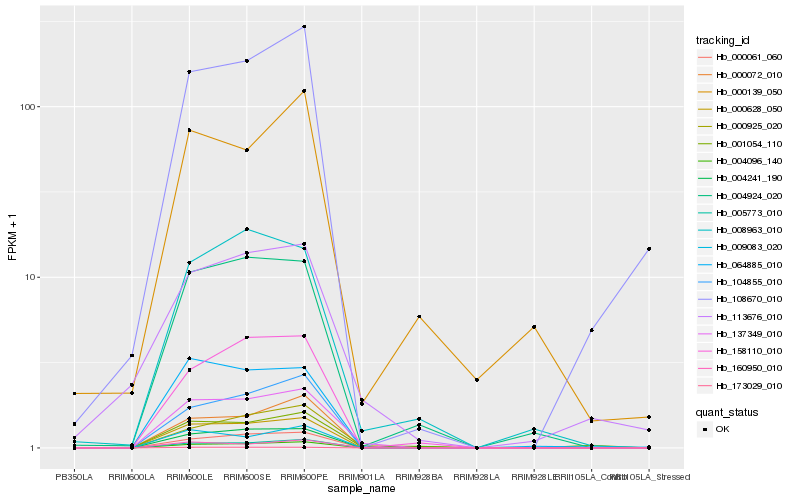
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001054_110 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 2 |
Hb_137349_010 |
0.1452997445 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 3 |
Hb_000925_020 |
0.1598449624 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 4 |
Hb_000628_050 |
0.1603131654 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_005773_010 |
0.1620259599 |
- |
- |
- |
| 6 |
Hb_004924_020 |
0.1662564358 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_064885_010 |
0.1700017232 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 8 |
Hb_160950_010 |
0.1713115684 |
- |
- |
PREDICTED: uncharacterized protein LOC105761043 [Gossypium raimondii] |
| 9 |
Hb_000072_010 |
0.1719840254 |
- |
- |
hypothetical protein B456_006G246700 [Gossypium raimondii] |
| 10 |
Hb_000139_050 |
0.1722280237 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 11 |
Hb_158110_010 |
0.1731002133 |
- |
- |
hypothetical protein CICLE_v10012017mg [Citrus clementina] |
| 12 |
Hb_108670_010 |
0.1731789317 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 13 |
Hb_004241_190 |
0.1757578718 |
- |
- |
- |
| 14 |
Hb_009083_020 |
0.1762514361 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 15 |
Hb_008963_010 |
0.1785569363 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_173029_010 |
0.1794393653 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_104855_010 |
0.1796264133 |
- |
- |
- |
| 18 |
Hb_000061_060 |
0.1800162984 |
transcription factor |
TF Family: MYB-related |
transcription factor MYB28 [Arabidopsis thaliana] |
| 19 |
Hb_113676_010 |
0.1823882549 |
- |
- |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 20 |
Hb_004096_140 |
0.1829602642 |
- |
- |
Lipase, putative [Ricinus communis] |