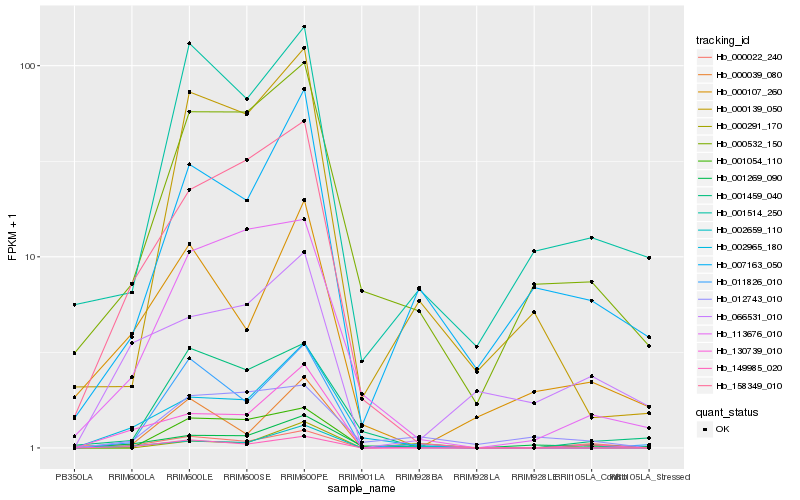
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000022_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638288 [Jatropha curcas] |
| 2 |
Hb_149985_020 |
0.0909213612 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Nicotiana tomentosiformis] |
| 3 |
Hb_001514_250 |
0.2409807346 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 4 |
Hb_001054_110 |
0.2447113918 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 5 |
Hb_000532_150 |
0.2449752807 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 6 |
Hb_007163_050 |
0.2548926363 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |
| 7 |
Hb_011826_010 |
0.2589444289 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 8 |
Hb_001269_090 |
0.2606838232 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 9 |
Hb_000107_260 |
0.2648488873 |
- |
- |
- |
| 10 |
Hb_002659_110 |
0.2661533399 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 11 |
Hb_066531_010 |
0.2704261862 |
- |
- |
ccd1, putative [Ricinus communis] |
| 12 |
Hb_113676_010 |
0.2719988343 |
- |
- |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 13 |
Hb_130739_010 |
0.2724002551 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 14 |
Hb_002965_180 |
0.2747615046 |
- |
- |
AMSH-like ubiquitin thioesterase 3 [Morus notabilis] |
| 15 |
Hb_000039_080 |
0.2787514268 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 16 |
Hb_000139_050 |
0.2808794792 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 17 |
Hb_012743_010 |
0.2809270399 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 18 |
Hb_158349_010 |
0.2815825694 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 19 |
Hb_001459_040 |
0.2824254052 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 20 |
Hb_000291_170 |
0.2836406501 |
- |
- |
PREDICTED: inactive rhomboid protein 1 isoform X1 [Jatropha curcas] |