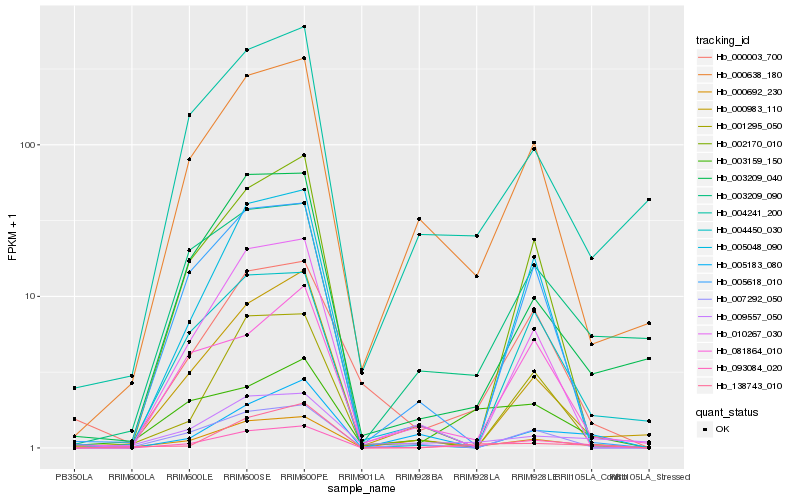
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_093084_020 |
0.0 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Populus euphratica] |
| 2 |
Hb_000638_180 |
0.1846225568 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 3 |
Hb_003209_090 |
0.2011873142 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| 4 |
Hb_004241_200 |
0.2035980826 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 5 |
Hb_000003_700 |
0.2040764868 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 6 |
Hb_009557_050 |
0.2093210898 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 7 |
Hb_003159_150 |
0.2101368283 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 8 [Jatropha curcas] |
| 8 |
Hb_007292_050 |
0.2128125784 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 9 |
Hb_004450_030 |
0.2142074385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003209_040 |
0.2160647063 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 11 |
Hb_005183_080 |
0.2165056603 |
- |
- |
- |
| 12 |
Hb_081864_010 |
0.219468765 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_005048_090 |
0.2211900864 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 14 |
Hb_138743_010 |
0.2219401716 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 15 |
Hb_005618_010 |
0.2222761696 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 16 |
Hb_000983_110 |
0.2226448998 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 17 |
Hb_001295_050 |
0.2233383627 |
- |
- |
PREDICTED: uncharacterized protein LOC101503108 isoform X1 [Cicer arietinum] |
| 18 |
Hb_002170_010 |
0.2234710243 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 19 |
Hb_010267_030 |
0.223557558 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000692_230 |
0.2246872346 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |