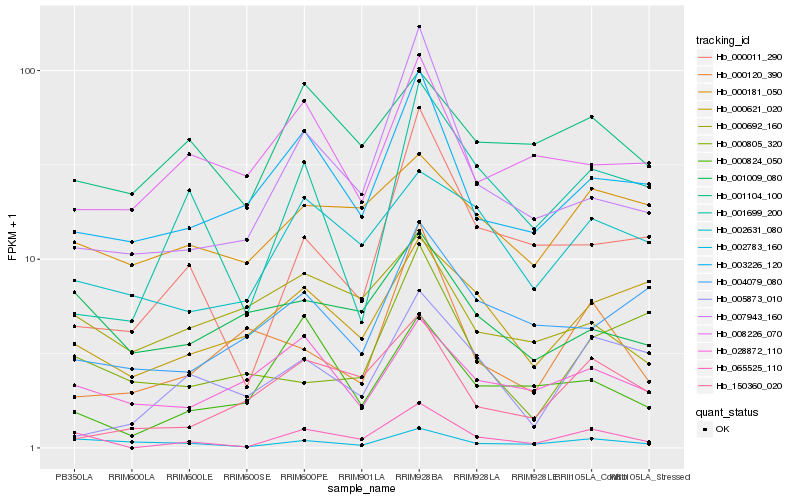
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065525_110 |
0.0 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-2 [Jatropha curcas] |
| 2 |
Hb_002783_160 |
0.2353678986 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 3 |
Hb_007943_160 |
0.235831627 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 4 |
Hb_003226_120 |
0.2473851384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000011_290 |
0.2486020712 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 6 |
Hb_000824_050 |
0.2584164142 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 7 |
Hb_001699_200 |
0.2610261034 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002631_080 |
0.2663314853 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_001009_080 |
0.2703280336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_028872_110 |
0.2742111874 |
- |
- |
PREDICTED: uncharacterized protein LOC105644776 [Jatropha curcas] |
| 11 |
Hb_000621_020 |
0.2745408478 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 12 |
Hb_001104_100 |
0.2805601929 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 13 |
Hb_150360_020 |
0.2817451642 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 14 |
Hb_000120_390 |
0.2849447428 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 15 |
Hb_000692_160 |
0.2866545781 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 16 |
Hb_000181_050 |
0.2868026464 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 17 |
Hb_008226_070 |
0.2900402832 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 18 |
Hb_000805_320 |
0.2904542408 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 19 |
Hb_004079_080 |
0.290820053 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 20 |
Hb_005873_010 |
0.2915487711 |
- |
- |
cysteine desulfurylase, putative [Ricinus communis] |