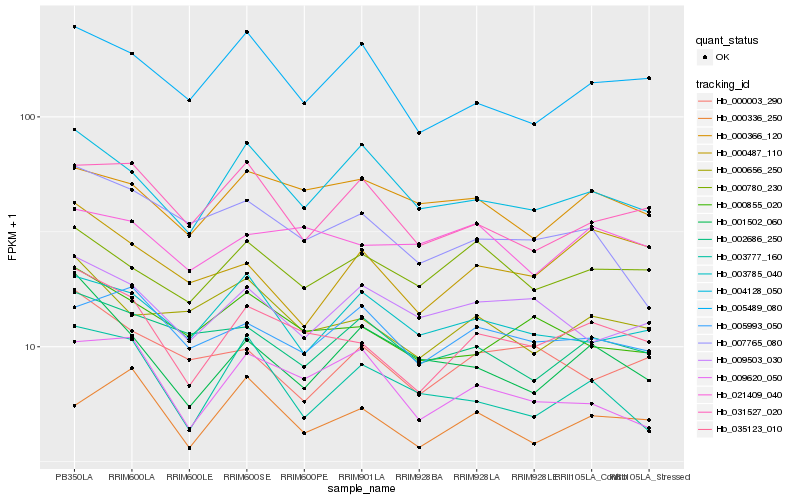
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_035123_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 2 |
Hb_000855_020 |
0.0924147346 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 3 |
Hb_000656_250 |
0.1003944135 |
- |
- |
hypothetical protein JCGZ_24680 [Jatropha curcas] |
| 4 |
Hb_005489_080 |
0.1032815139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_009620_050 |
0.1057177318 |
- |
- |
PREDICTED: protein unc-45 homolog A [Jatropha curcas] |
| 6 |
Hb_000780_230 |
0.1058498304 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 7 |
Hb_004128_050 |
0.1060411593 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_021409_040 |
0.109130424 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 9 |
Hb_000366_120 |
0.1096762487 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 10 |
Hb_000336_250 |
0.1103003529 |
transcription factor |
TF Family: mTERF |
hypothetical protein B456_002G027700 [Gossypium raimondii] |
| 11 |
Hb_031527_020 |
0.1108057187 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 12 |
Hb_005993_050 |
0.1115648257 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 13 |
Hb_000487_110 |
0.1119232745 |
- |
- |
PREDICTED: cyclin-T1-3-like [Jatropha curcas] |
| 14 |
Hb_000003_290 |
0.1132785042 |
- |
- |
hypothetical protein JCGZ_24486 [Jatropha curcas] |
| 15 |
Hb_007765_080 |
0.1134829183 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 16 |
Hb_009503_030 |
0.114349531 |
- |
- |
protein dimerization, putative [Ricinus communis] |
| 17 |
Hb_003777_160 |
0.1149275069 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001502_060 |
0.114965084 |
- |
- |
DNA mismatch repair protein MSH2, putative [Ricinus communis] |
| 19 |
Hb_002686_250 |
0.1150479009 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 20 |
Hb_003785_040 |
0.1152585959 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |