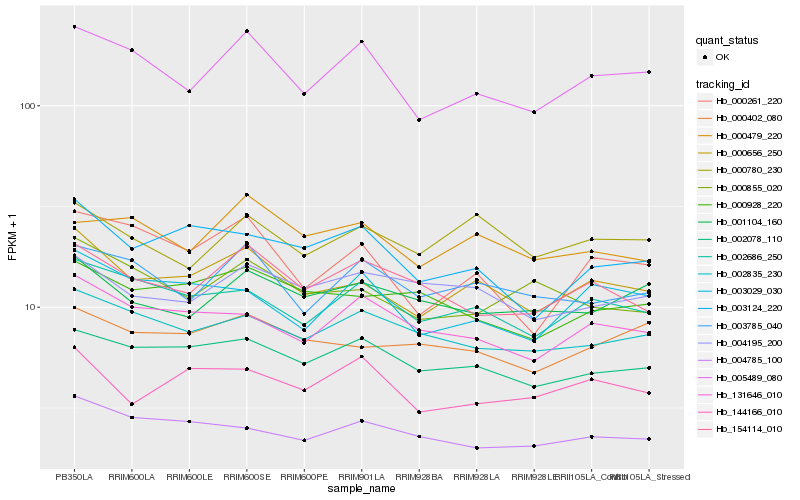
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_250 |
0.0 |
- |
- |
hypothetical protein JCGZ_24680 [Jatropha curcas] |
| 2 |
Hb_002686_250 |
0.0732113107 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 3 |
Hb_002078_110 |
0.0782390436 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 4 |
Hb_005489_080 |
0.0810780543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000402_080 |
0.0817431313 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 6 |
Hb_000780_230 |
0.0851928247 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 7 |
Hb_131646_010 |
0.0867585669 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_003124_220 |
0.0869343314 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 9 |
Hb_002835_230 |
0.0873434785 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000928_220 |
0.0877184332 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 11 |
Hb_000261_220 |
0.0882530652 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 12 |
Hb_000855_020 |
0.0895506589 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 13 |
Hb_004785_100 |
0.0899114851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003785_040 |
0.0904647148 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_003029_030 |
0.0906580214 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004195_200 |
0.091756688 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 17 |
Hb_144166_010 |
0.0921781277 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 18 |
Hb_154114_010 |
0.0928824405 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 19 |
Hb_001104_160 |
0.0938883087 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 20 |
Hb_000479_220 |
0.096218987 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |