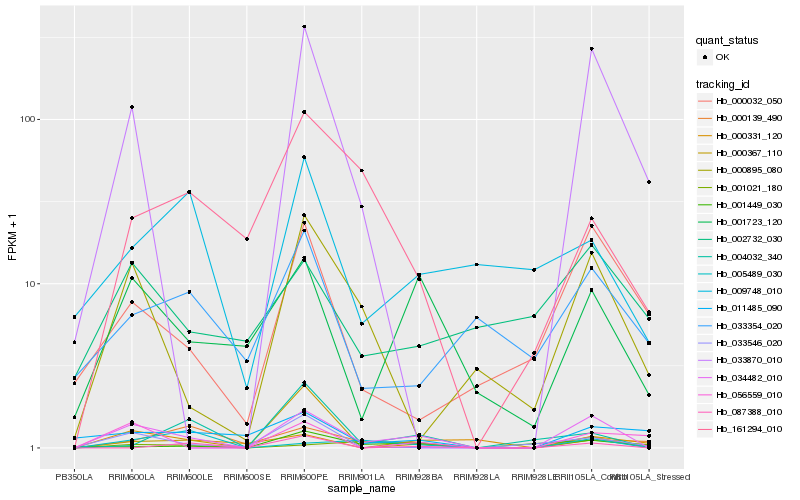
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034482_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001723_120 |
0.3136407857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000895_080 |
0.3277276642 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_033870_010 |
0.342748571 |
- |
- |
PREDICTED: uncharacterized protein LOC104101109 isoform X1 [Nicotiana tomentosiformis] |
| 5 |
Hb_033546_020 |
0.3561167095 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 6 |
Hb_000032_050 |
0.3605920597 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |
| 7 |
Hb_056559_010 |
0.3816913673 |
- |
- |
PREDICTED: uncharacterized protein LOC104088595 [Nicotiana tomentosiformis] |
| 8 |
Hb_161294_010 |
0.3834378976 |
- |
- |
PREDICTED: serine/threonine-protein kinase ppk15 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001449_030 |
0.3926475089 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000367_110 |
0.3949392797 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic isoform X1 [Sesamum indicum] |
| 11 |
Hb_087388_010 |
0.396273392 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 12 |
Hb_001021_180 |
0.400457095 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Nicotiana tomentosiformis] |
| 13 |
Hb_033354_020 |
0.4037252634 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_011485_090 |
0.4038858967 |
- |
- |
- |
| 15 |
Hb_000331_120 |
0.4048339216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_009748_010 |
0.4048402064 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 17 |
Hb_000139_490 |
0.4052583001 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 18 |
Hb_002732_030 |
0.4063425207 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005489_030 |
0.4072204391 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 20 |
Hb_004032_340 |
0.4085089977 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |