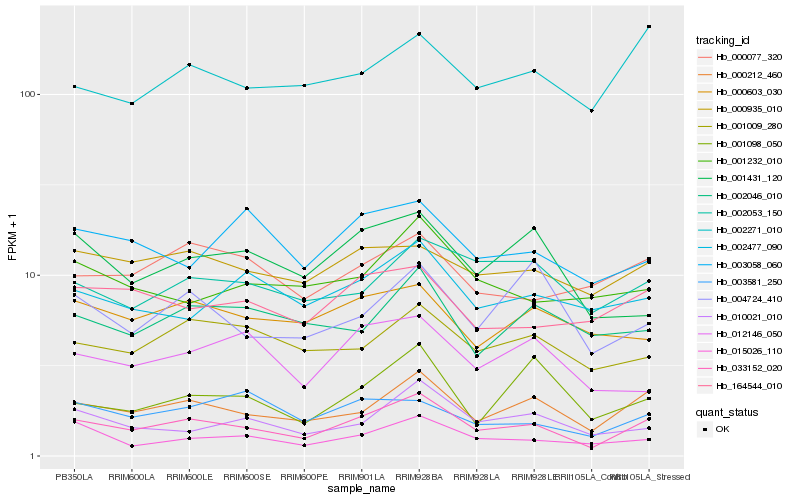
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033152_020 |
0.0 |
- |
- |
hypothetical protein VITISV_000649 [Vitis vinifera] |
| 2 |
Hb_000212_460 |
0.0943275505 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 3 |
Hb_015026_110 |
0.1411499502 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_010021_010 |
0.1425015036 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_002271_010 |
0.147861694 |
- |
- |
PREDICTED: 60S ribosomal protein L8 [Jatropha curcas] |
| 6 |
Hb_001009_280 |
0.1485827306 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 7 |
Hb_002053_150 |
0.1499866888 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 8 |
Hb_000935_010 |
0.1502807964 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 9 |
Hb_164544_010 |
0.1506470203 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.5 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000077_320 |
0.154180382 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 11 |
Hb_004724_410 |
0.1542210825 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002477_090 |
0.1546101435 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001232_010 |
0.154733316 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 14 |
Hb_001431_120 |
0.1553138021 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 15 |
Hb_012146_050 |
0.1566239857 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002046_010 |
0.1580713453 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003058_060 |
0.1589923459 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 18 |
Hb_000603_030 |
0.1604822903 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 19 |
Hb_003581_250 |
0.1604847824 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 20 |
Hb_001098_050 |
0.1606701586 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |