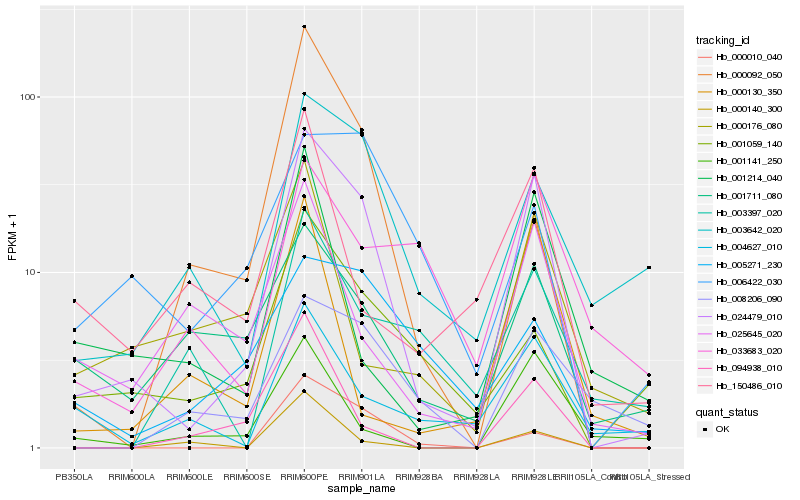
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024479_010 |
0.0 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 2 |
Hb_000010_040 |
0.2576108338 |
- |
- |
PREDICTED: probable 1-aminocyclopropane-1-carboxylate deaminase-like isoform X2 [Citrus sinensis] |
| 3 |
Hb_003642_020 |
0.2616965619 |
- |
- |
chlorophyll(ide) b reductase, partial [Pyrus x bretschneideri] |
| 4 |
Hb_001141_250 |
0.2818751512 |
- |
- |
hypothetical protein glysoja_047929, partial [Glycine soja] |
| 5 |
Hb_008206_090 |
0.2822630548 |
- |
- |
PREDICTED: uncharacterized protein LOC100811033 [Glycine max] |
| 6 |
Hb_094938_010 |
0.2833207755 |
- |
- |
- |
| 7 |
Hb_001059_140 |
0.2877631753 |
- |
- |
hypothetical protein POPTR_0004s03390g [Populus trichocarpa] |
| 8 |
Hb_001711_080 |
0.2897419622 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 9 |
Hb_025645_020 |
0.2924366165 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 10 |
Hb_001214_040 |
0.2924708467 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 11 |
Hb_000092_050 |
0.2925016782 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 12 |
Hb_004627_010 |
0.2936182255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000140_300 |
0.306615784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006422_030 |
0.3070102599 |
- |
- |
- |
| 15 |
Hb_150486_010 |
0.3077982883 |
- |
- |
hypothetical protein CICLE_v100116733mg, partial [Citrus clementina] |
| 16 |
Hb_005271_230 |
0.3081204835 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X4 [Jatropha curcas] |
| 17 |
Hb_033683_020 |
0.3089079139 |
- |
- |
hypothetical protein POPTR_1072s00200g [Populus trichocarpa] |
| 18 |
Hb_000130_350 |
0.3144515709 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 19 |
Hb_000176_080 |
0.3170634157 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |
| 20 |
Hb_003397_020 |
0.3188195061 |
- |
- |
- |