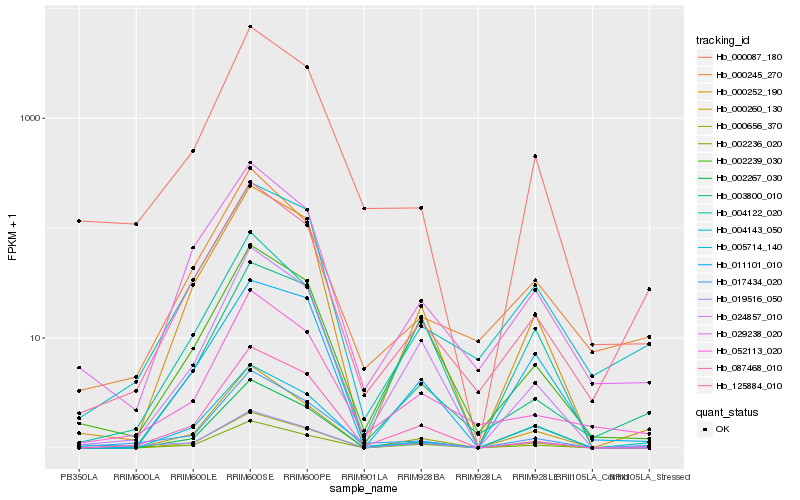
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019516_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_04619 [Jatropha curcas] |
| 2 |
Hb_017434_020 |
0.1397887785 |
transcription factor |
TF Family: C2H2 |
Zinc finger protein 4 [Populus trichocarpa] |
| 3 |
Hb_004143_050 |
0.1461324418 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 4 |
Hb_000252_190 |
0.148583091 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 5 |
Hb_002236_020 |
0.1506536667 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 6 |
Hb_004122_020 |
0.1535583221 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 7 |
Hb_011101_010 |
0.1621044048 |
- |
- |
hypothetical protein JCGZ_25132 [Jatropha curcas] |
| 8 |
Hb_024857_010 |
0.1641134169 |
- |
- |
glutaredoxin 2.1 [Hevea brasiliensis] |
| 9 |
Hb_002267_030 |
0.1682837809 |
- |
- |
hypothetical protein JCGZ_24883 [Jatropha curcas] |
| 10 |
Hb_000656_370 |
0.1691855569 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_125884_010 |
0.1715998799 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Jatropha curcas] |
| 12 |
Hb_000245_270 |
0.1739942196 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 13 |
Hb_005714_140 |
0.1743607818 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_003800_010 |
0.1753108248 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 15 |
Hb_029238_020 |
0.1760672424 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 16 |
Hb_002239_030 |
0.1761616116 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 17 |
Hb_000087_180 |
0.1781666339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000260_130 |
0.1788093141 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_087468_010 |
0.1788954858 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 20 |
Hb_052113_020 |
0.1789996043 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |