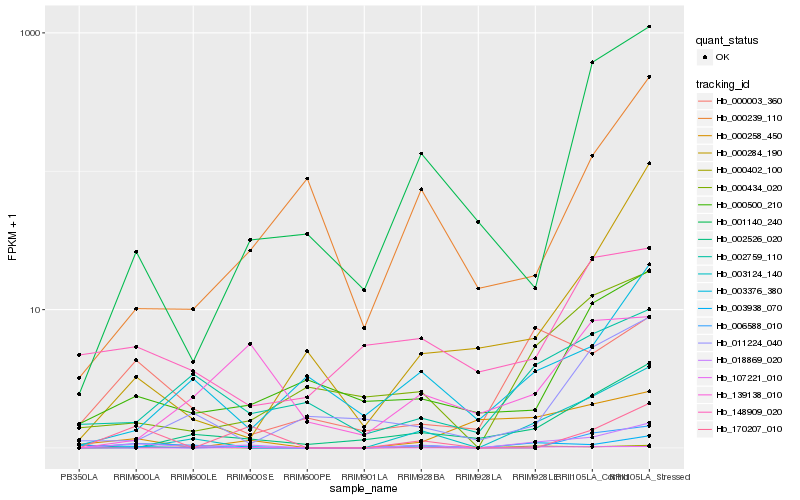
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018869_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 2 |
Hb_003124_140 |
0.2554872148 |
- |
- |
hypothetical protein Csa_4G182240 [Cucumis sativus] |
| 3 |
Hb_000434_020 |
0.2779853154 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 4 |
Hb_003938_070 |
0.2830260397 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_002526_020 |
0.2846059003 |
- |
- |
- |
| 6 |
Hb_002759_110 |
0.292735808 |
- |
- |
RNA-binding family protein, putative isoform 1 [Theobroma cacao] |
| 7 |
Hb_000500_210 |
0.3013005277 |
- |
- |
- |
| 8 |
Hb_006588_010 |
0.3015699997 |
- |
- |
- |
| 9 |
Hb_107221_010 |
0.3064636112 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_003376_380 |
0.3120894268 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 11 |
Hb_000003_360 |
0.316960257 |
- |
- |
Putative inactive purple acid phosphatase 27 [Glycine soja] |
| 12 |
Hb_000284_190 |
0.3169815997 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 13 |
Hb_001140_240 |
0.3179261138 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_000402_100 |
0.3227976603 |
- |
- |
PREDICTED: L-tryptophan--pyruvate aminotransferase 1 [Jatropha curcas] |
| 15 |
Hb_000258_450 |
0.3310606042 |
- |
- |
- |
| 16 |
Hb_011224_040 |
0.3354292625 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 17 |
Hb_170207_010 |
0.3387227998 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 18 |
Hb_148909_020 |
0.3409584109 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_000239_110 |
0.3417483756 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_139138_010 |
0.3449789704 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |