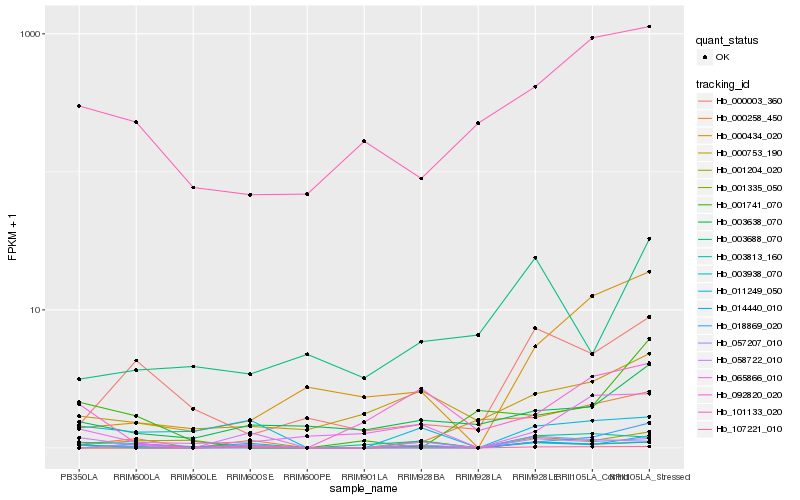
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003938_070 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_018869_020 |
0.2830260397 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 3 |
Hb_003813_160 |
0.3100246753 |
- |
- |
- |
| 4 |
Hb_003638_070 |
0.3184718448 |
- |
- |
hypothetical protein EUGRSUZ_E00638 [Eucalyptus grandis] |
| 5 |
Hb_000753_190 |
0.3303961582 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Populus euphratica] |
| 6 |
Hb_000003_360 |
0.3360701167 |
- |
- |
Putative inactive purple acid phosphatase 27 [Glycine soja] |
| 7 |
Hb_001204_020 |
0.3424357715 |
- |
- |
Uncharacterized protein TCM_021965 [Theobroma cacao] |
| 8 |
Hb_107221_010 |
0.3448709204 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_014440_010 |
0.3463160668 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_001335_050 |
0.3476985355 |
- |
- |
- |
| 11 |
Hb_101133_020 |
0.3477514884 |
- |
- |
PREDICTED: 15.7 kDa heat shock protein, peroxisomal [Jatropha curcas] |
| 12 |
Hb_000434_020 |
0.3489362824 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 13 |
Hb_003688_070 |
0.3505551703 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein E [Jatropha curcas] |
| 14 |
Hb_011249_050 |
0.3519810366 |
- |
- |
- |
| 15 |
Hb_092820_020 |
0.3538142762 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000258_450 |
0.3540960373 |
- |
- |
- |
| 17 |
Hb_065866_010 |
0.3565753742 |
- |
- |
PREDICTED: hydroxyacyl-thioester dehydratase type 2, mitochondrial [Fragaria vesca subsp. vesca] |
| 18 |
Hb_058722_010 |
0.3607575637 |
- |
- |
- |
| 19 |
Hb_057207_010 |
0.3612900513 |
- |
- |
hypothetical protein F383_17010 [Gossypium arboreum] |
| 20 |
Hb_001741_070 |
0.368316047 |
rubber biosynthesis |
Gene Name: SRPP7 |
unnamed protein product [Coffea canephora] |