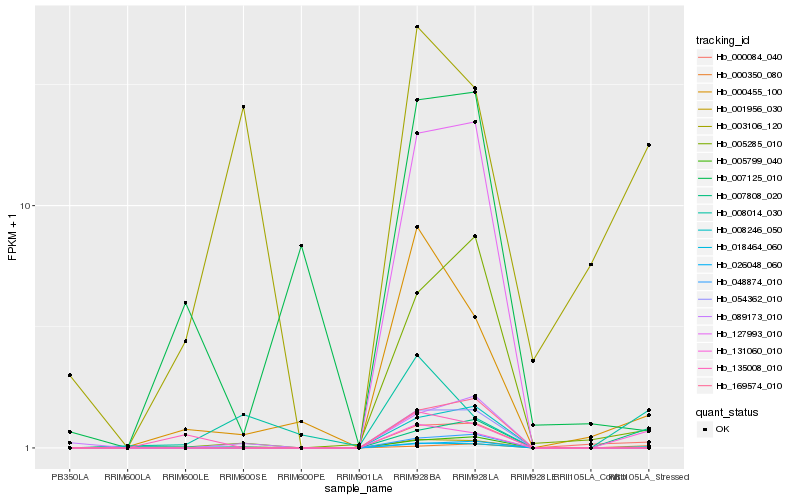
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018464_060 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_000455_100 |
0.2712890423 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_001956_030 |
0.2728263898 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Populus euphratica] |
| 4 |
Hb_000084_040 |
0.2758270975 |
- |
- |
hypothetical protein RCOM_1007940 [Ricinus communis] |
| 5 |
Hb_003106_120 |
0.284355015 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 6 |
Hb_131060_010 |
0.3004264588 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 7 |
Hb_005285_010 |
0.3009334002 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_007808_020 |
0.312028474 |
- |
- |
- |
| 9 |
Hb_026048_060 |
0.3172030875 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 10 |
Hb_054362_010 |
0.3175937399 |
- |
- |
PREDICTED: uncharacterized protein LOC105650428 [Jatropha curcas] |
| 11 |
Hb_127993_010 |
0.3201806871 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 12 |
Hb_135008_010 |
0.3269906526 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 13 |
Hb_000350_080 |
0.3272979492 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 14 |
Hb_169574_010 |
0.3299995073 |
- |
- |
PREDICTED: uncharacterized protein LOC104120373 [Nicotiana tomentosiformis] |
| 15 |
Hb_048874_010 |
0.330691712 |
- |
- |
hypothetical protein POPTR_0016s10870g [Populus trichocarpa] |
| 16 |
Hb_005799_040 |
0.3324665478 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4-like isoform X1 [Gossypium raimondii] |
| 17 |
Hb_008246_050 |
0.3324799588 |
- |
- |
- |
| 18 |
Hb_089173_010 |
0.3333054282 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 19 |
Hb_008014_030 |
0.3350668731 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 20 |
Hb_007125_010 |
0.3351392736 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |