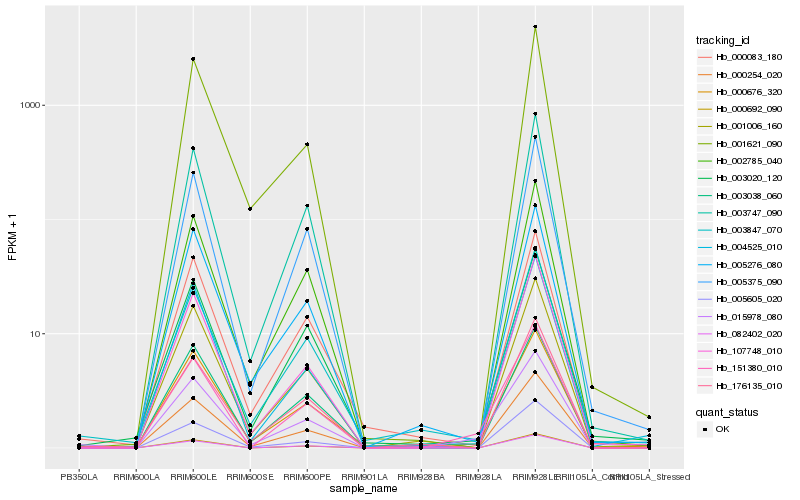
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015978_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_151380_010 |
0.0821239496 |
- |
- |
ribosomal protein S4 [Hevea brasiliensis] |
| 3 |
Hb_000676_320 |
0.1117191308 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 4 |
Hb_003847_070 |
0.1175947439 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003038_060 |
0.1213311354 |
- |
- |
hypothetical protein POPTR_0007s13050g [Populus trichocarpa] |
| 6 |
Hb_000254_020 |
0.1223386394 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 7 |
Hb_003747_090 |
0.1224778375 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_002785_040 |
0.1226948974 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 9 |
Hb_000083_180 |
0.1254110682 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 10 |
Hb_176135_010 |
0.1261389689 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_005276_080 |
0.1267323023 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005375_090 |
0.1268328132 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001006_160 |
0.1287302449 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_000692_090 |
0.1291403564 |
- |
- |
- |
| 15 |
Hb_005605_020 |
0.1291615927 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 16 |
Hb_107748_010 |
0.1292662502 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_003020_120 |
0.1293980689 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 18 |
Hb_001621_090 |
0.129549203 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 19 |
Hb_082402_020 |
0.1302397277 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 20 |
Hb_004525_010 |
0.1304096324 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |