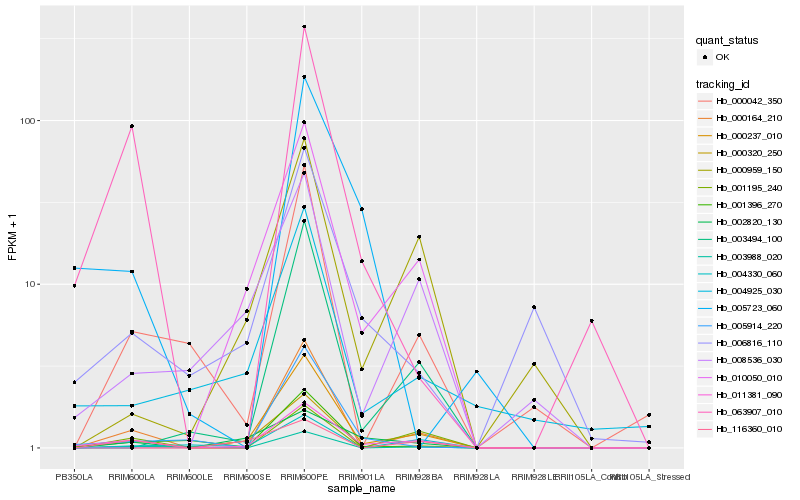
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011381_090 |
0.0 |
- |
- |
Protein ABC1, mitochondrial precursor, putative [Ricinus communis] |
| 2 |
Hb_000164_210 |
0.2279784123 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 3 |
Hb_063907_010 |
0.2503338461 |
- |
- |
- |
| 4 |
Hb_001195_240 |
0.2557867676 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein RADIALIS-like 3 [Jatropha curcas] |
| 5 |
Hb_002820_130 |
0.2633332936 |
- |
- |
- |
| 6 |
Hb_008536_030 |
0.2634885477 |
- |
- |
hypothetical protein POPTR_0005s08290g [Populus trichocarpa] |
| 7 |
Hb_000237_010 |
0.2653861004 |
transcription factor |
TF Family: SRS |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006816_110 |
0.2674052329 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 1 [Jatropha curcas] |
| 9 |
Hb_010050_010 |
0.2687469076 |
- |
- |
- |
| 10 |
Hb_116360_010 |
0.2706838375 |
- |
- |
- |
| 11 |
Hb_004925_030 |
0.2735929202 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 12 |
Hb_000959_150 |
0.2770030634 |
- |
- |
PREDICTED: uncharacterized protein LOC104879927 [Vitis vinifera] |
| 13 |
Hb_003988_020 |
0.2776132859 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH85-like [Populus euphratica] |
| 14 |
Hb_004330_060 |
0.2778993336 |
- |
- |
hypothetical protein POPTR_0011s01530g [Populus trichocarpa] |
| 15 |
Hb_000042_350 |
0.2789644261 |
- |
- |
hypothetical protein JCGZ_00710 [Jatropha curcas] |
| 16 |
Hb_001396_270 |
0.2821167793 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 17 |
Hb_000320_250 |
0.2840068482 |
- |
- |
hypothetical protein POPTR_0021s00440g [Populus trichocarpa] |
| 18 |
Hb_005723_060 |
0.2873149847 |
- |
- |
- |
| 19 |
Hb_003494_100 |
0.2882137979 |
transcription factor |
TF Family: zf-HD |
Mini zinc finger 2 isoform 1 [Theobroma cacao] |
| 20 |
Hb_005914_220 |
0.2884998724 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |