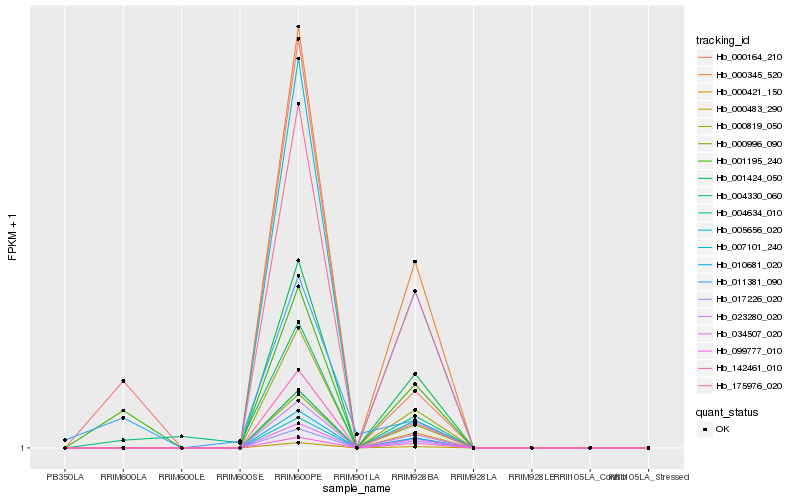
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_240 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein RADIALIS-like 3 [Jatropha curcas] |
| 2 |
Hb_000164_210 |
0.2246263188 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 3 |
Hb_004330_060 |
0.2469734919 |
- |
- |
hypothetical protein POPTR_0011s01530g [Populus trichocarpa] |
| 4 |
Hb_005656_020 |
0.2508874544 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 5 |
Hb_001424_050 |
0.2509128675 |
- |
- |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_175976_020 |
0.2510978239 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 7 |
Hb_034507_020 |
0.2511098869 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 8 |
Hb_017226_020 |
0.2511612898 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_06720 [Jatropha curcas] |
| 9 |
Hb_142461_010 |
0.2515377229 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_000819_050 |
0.2528342538 |
- |
- |
hypothetical protein Csa_4G290160 [Cucumis sativus] |
| 11 |
Hb_023280_020 |
0.2532157561 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 12 |
Hb_000345_520 |
0.2533982767 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |
| 13 |
Hb_000996_090 |
0.2542191067 |
transcription factor |
TF Family: NF-YC |
ccaat-binding transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000483_290 |
0.2553796549 |
- |
- |
PREDICTED: ankyrin-1-like [Prunus mume] |
| 15 |
Hb_010681_020 |
0.2554217472 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 16 |
Hb_007101_240 |
0.2556079312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_011381_090 |
0.2557867676 |
- |
- |
Protein ABC1, mitochondrial precursor, putative [Ricinus communis] |
| 18 |
Hb_000421_150 |
0.2558292139 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 19 |
Hb_099777_010 |
0.2561516911 |
- |
- |
PREDICTED: uncharacterized protein LOC104884611 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_004634_010 |
0.2574627569 |
- |
- |
hypothetical protein POPTR_0002s23100g [Populus trichocarpa] |