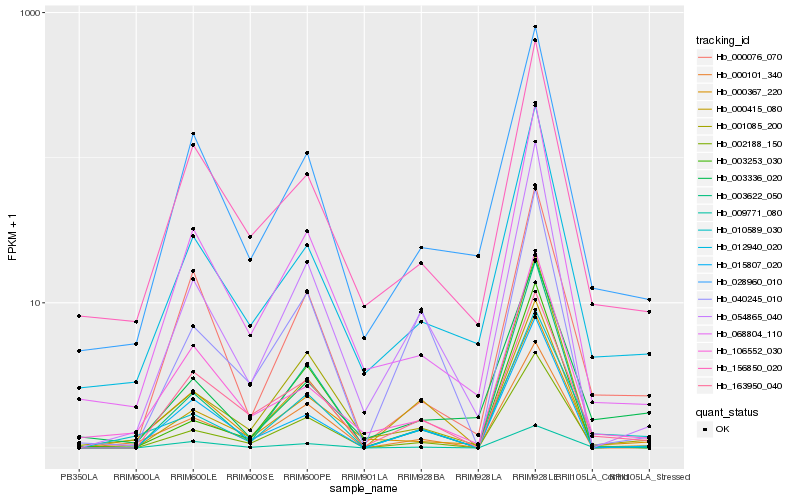
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010589_030 |
0.0 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 2 |
Hb_001085_200 |
0.1464891238 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 3 |
Hb_002188_150 |
0.1531765481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012940_020 |
0.1593944148 |
- |
- |
zeaxanthin epoxidase, putative [Ricinus communis] |
| 5 |
Hb_000101_340 |
0.1619455846 |
- |
- |
2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Ricinus communis] |
| 6 |
Hb_054865_040 |
0.1682903825 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 7 |
Hb_009771_080 |
0.1710651033 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 8 |
Hb_003336_020 |
0.1736020935 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS3, chloroplastic [Vitis vinifera] |
| 9 |
Hb_028960_010 |
0.1739719099 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 10 |
Hb_000367_220 |
0.1760459168 |
- |
- |
hypothetical protein RCOM_1470580 [Ricinus communis] |
| 11 |
Hb_000076_070 |
0.1760553399 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 12 |
Hb_068804_110 |
0.176847467 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000415_080 |
0.176953639 |
- |
- |
- |
| 14 |
Hb_003622_050 |
0.1790919918 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A-like [Jatropha curcas] |
| 15 |
Hb_163950_040 |
0.1803164044 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003253_030 |
0.1810820207 |
- |
- |
lysosomal pro-X carboxypeptidase, putative [Ricinus communis] |
| 17 |
Hb_156850_020 |
0.1843340539 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |
| 18 |
Hb_015807_020 |
0.1868258901 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 19 |
Hb_106552_030 |
0.1912895752 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 20 |
Hb_040245_010 |
0.1921836591 |
- |
- |
hypothetical protein CISIN_1g039575mg, partial [Citrus sinensis] |